Figures & data
Figure 1. Validation of the phosphotyrosine (PY) and RTK immunoaffinity double purification approach in GIST882 by immunoblot. Total protein lysate (input) is included as a control. KIT, panRTK, PY, and RTK-sepharose IPs are positive controls. Pre-immune IP is a negative control. PY staining after double purification (PY immunoaffinity column followed by panRTK IP) shows a doublet representing the known KIT oncoprotein. The presence of KIT was corroborated by mass spectrometry analysis.
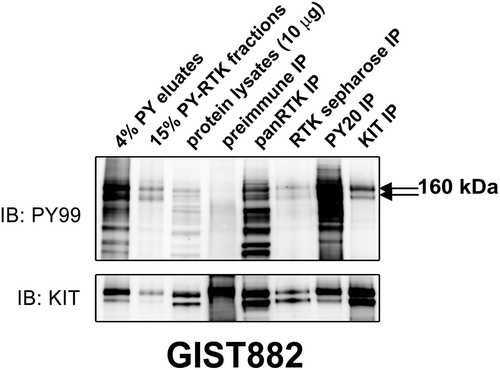
Table 1. Identification of novel candidate protein tyrosine kinase oncoproteins in KIT-negative cell lines (GIST62 and GIST522) by the phosphotyrosine-RTK immuno-affinity purification and tandem mass spectrometry.
Figure 2. Phosphorylation of purified activated tyrosine kinases in KIT-negative GIST62 and GIST522 were evaluated by staining with PY and specific antibodies (AXL, FAK, EGFR, PDGFRA, KIT, EPHA2, and paxillin) in immunoblots. Total protein lysate (input) is a total protein control. PY and total staining of PY eluates and PY-RTK fractions reveal bands corresponding to AXL and FAK, which were corroborated by mass spectrometry analysis.
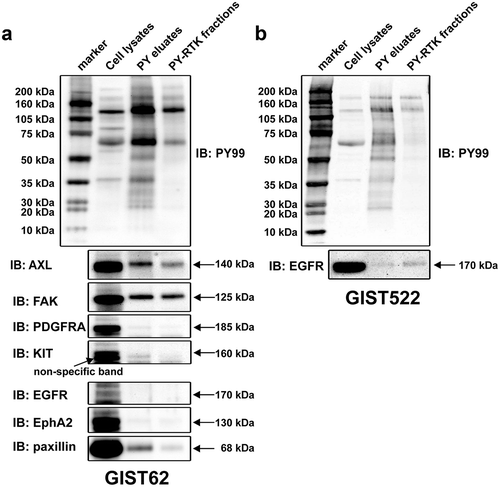
Figure 3. (a) Immunoblotting evaluations of the relative expression and phosphorylation levels of candidate tyrosine kinase oncoproteins including FAK, AXL, EPHA2, EGFR, PDGFRA, and MET, between KIT-negative GISTs (GIST62 and GIST522), and KIT-positive GISTs (GIST882, GIST48, and GIST430). MESO257, RMS556, and MCF-7 are included as controls. Actin staining is a loading control. (b) Phosphorylation levels of AXL, EPHA2, FAK, and EGFR were evaluated by immunoprecipitation followed by PY and specific antibody immunoblotting in GIST62 and GIST522. Total protein lysate is a total protein control. (c) Quantitative RT-PCR evaluations of AXL transcript expression in GIST cell lines. AXL expression was greater in KIT-negative GIST62 and GIST522 than in KIT-positive GIST882, GIST48, and GIST430. AXL expression was induced after imatinib selection in GIST882IMR, which has lost KIT oncogene expression, as compared to parental GIST882. (d) Immunoblotting evaluations of the expression of AXL and KIT in KIT-positive or -negative frozen clinical GISTs. Actin staining is a loading control.
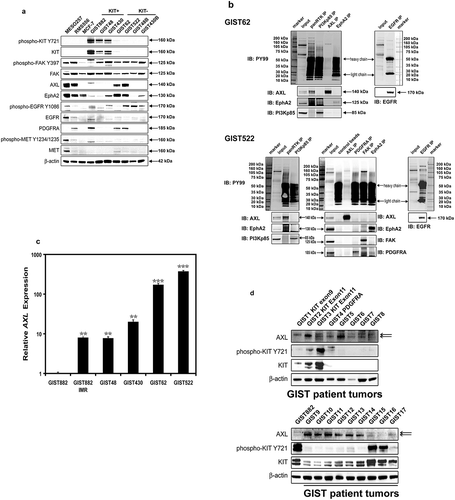
Figure 4. (a) Immunoblotting evaluations of GIST522 and GIST48B after treatment with gefitinib for 4h in serum-free media. MESO924 is an EGFR-positive control, GIST882 is an EGFR-negative control. (b) Cell viability was evaluated in GIST522 (black bars), GIST48B (light gray bars), and PC-9 (dark gray bars), at day 6 after treatment with gefitinib. Viability was analyzed using Cell-titer Glo® ATP-based luminescence assay. The data were normalized to DMSO and represent the mean values (± s.d.) from quadruplicate cultures. Statistically significant differences between DMSO control and gefitinib treatments are presented as *p < 0.05, ***p < 0.001. (c) Colony growth assays were performed 10 days after treatment with gefitinib (100, 250, 500, 1000, and 2500 nM). EGFR inhibition led to a greater reduction in colony formation and size in PC-9, but not in GIST48B. (d) Quantitations of GIST48B and PC-9 cell colony growth after treatment with gefitinib for 10 days. Statistically significant differences between DMSO control and inhibitor treatments are presented as ***p < 0.001.
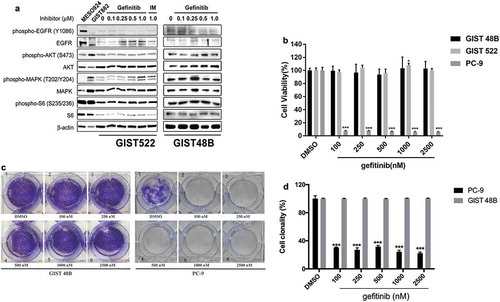
Figure 5. (a) Immunoblotting evaluations of KIT-negative GIST cell lines (GIST62 and GIST522) and KIT-positive GIST cell line (GIST882) at 96 hours after infection by lentiviral AXL, EPHA2, and FAK shRNA constructs. Immunoblots demonstrate the effects of AXL, EPHA2 and FAK knockdown on signaling intermediates (AKT, MAPK p42/44, S6, STAT1, and STAT3), proliferation markers (Cyclin A and PCNA), and cell cycle checkpoint proteins (p27, p21, and p53). Control lanes for each cell line include uninfected cells (untreated lane) and cells infected with empty lentiviral vector. Actin is a loading control. (b) Immunoblotting evaluations of GIST522 and GIST882 cells at 10 days after infection by lentiviral AXL, EPHA2, and FAK shRNA constructs. AXL shRNA resulted in decreased expression of cyclin A and PCNA, and upregulation of p53, p21, and p27 in GIST522, but not in GIST882. Actin is a loading control. (c) GIST62 and GIST522 cell cultures, evaluated at 6 days after infection by lentiviral AXL shRNA constructs, showing growth inhibition as compared to control cultures infected with empty lentiviral constructs. Scale bars: 100 μm. (d) Cell viability was evaluated in GIST62 (black bars), GIST522 (white bars), and GIST430 (gray bars), at day 8 and day 12 after infection with lentiviral AXL, EPHA2, or FAK shRNA. Viability was analyzed using the Cell-titer Glo® ATP-based luminescence assay. The data were normalized to empty lentiviral infections, and represent the mean values (± s.d.) from quadruplicate cultures. The experiments were performed in triplicate, and statistically significant differences between vector control and shRNAs are defined as *p < 0.05, ** p < 0.01, *** p < 0.001. (e) Cell cycle analyses were performed in GIST62 cells at 4 days after infection by lentiviral AXL shRNA constructs. GIST62 cells showed a G2 block following AXL silencing as compared to empty vector control. Cells treated with AXL shRNA showed increased nuclear fragmentation (pre-G1), the experiments were performed in triplicate.
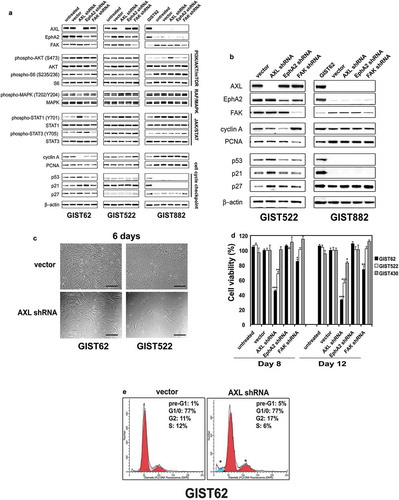
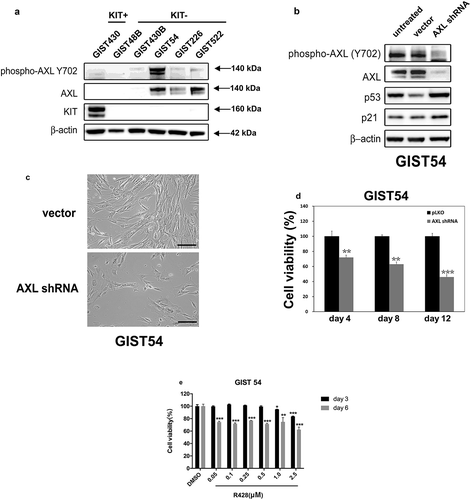
Figure 6. (a) Immunoblotting evaluations of the expression and phosphorylation of AXL between KIT-negative GISTs (GIST48B, GIST430B, GIST62, GIST226, and GIST522), and KIT-positive GISTs (GIST430). Actin staining is a loading control. (b) Immunoblots demonstrate the effects of AXL knockdown on expression of p53 and p21. (c) GIST54 cell cultures, evaluated at 6 days after infection by lentiviral AXL shRNA constructs, showing growth inhibition as compared to control cultures infected with empty lentiviral constructs. Scale bars: 100 μm. (d) Cell viability was evaluated in GIST54, at day 4, day 8, and day 12 after infection with lentiviral AXL. Viability was analyzed using the Cell-titer Glo® ATP-based luminescence assay. The data were normalized to empty lentiviral infections, and represent the mean values (± s.d.) from quadruplicate cultures. The experiments were performed in triplicate, and statistically significant differences between vector control and shRNAs are defined as ** p < 0.01, *** p < 0.001. (e) Cell viability was evaluated in GIST54, at day 3 (black bars) and day 6 (gray bars) after treatment with R428. Viability was analyzed using Cell-titer Glo® ATP-based luminescence assay. The data were normalized to DMSO and represent the mean values (± s.d.) from quadruplicate cultures. Statistically significant differences between DMSO control and R428 treatments are presented as *p < 0.05, ** p < 0.01, *** p < 0.001.