Figures & data
Table 1. HORMA-containing proteins from taxonomically representative eukaryotic and prokaryotic organisms. HORMA-containing proteins from human, yeast and Arabidopsis were used as query sequences in NCBI BlastP and UniProt to retrieve HORMA-containing proteins in eukaryotes and prokaryotes. Twenty-nine eukaryotic proteins were selected from NCBI and 22 prokaryotic proteins were selected from UniPro for phylogenetic analysis and investigation of HORMA-containing proteins structure and architecture (Accession numbers shown in the 3rd column).
Figure 1. Phylogenetic phenogram tree produced from the alignment of 29 eukaryotic and 22 prokaryotic HORMA-containing proteins. Multiple sequence alignment was constructed by MUSCLE 3.8 and phylogenetic trees generated using MEGA 7.0.26 software. (a) Phylogenetic tree constructed by the NJ method, (b) Phylogenetic tree constructed by ML method. Numbers on nodes are bootstrap percentages supporting a given partitioning. The proteins are designated by the genus name followed by the name of protein. Uncharacterized proteins are named by genus name followed by “HORMA”. Streptomyces purpurogeneiscleroticus # A0A0M8ZCY4 named as “StreptomycesHORMA”, Streptomyces sp. # A0A2A2Z569 named as “StreptomycesHORMA1”. Accession numbers for all proteins are listed in . Red, blue and green circles indicate for eukaryotic, bacterial and archaeal HORMA-containing proteins, respectively.
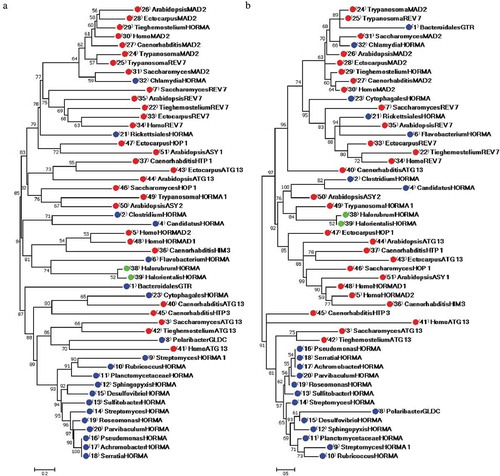
Figure 2. HORMA domain architecture in some HORMA-containing proteins from (a) eukaryotes and (b) prokaryotes. In eukaryotes, HORMA domain size ranged from 177 to 264 aa. Prokaryotic HORMA-containing proteins were classified based on domain size and multiple sequence alignment results into three groups. HORMA-containing proteins in the first group ranged in size from 165 to 167 aa and HORMA domains in these group ranged from 148–159 aa. The second group contained larger HORMA domains (from 196 to 224 aa), while the third group contained smaller HORMA domains (from 52 to 145 aa). ScanProsite search used to retrieve domain architecture in all eukaryotic HORMA-containing proteins and three bacterial proteins; Chlamydia trachomatis, Cytophagales bacterium and Rickettsiales bacterium. HORMA domains architecture in other prokaryotic proteins were drawn using MyDomains- Image Creator based on proteins structure information in InterPro.
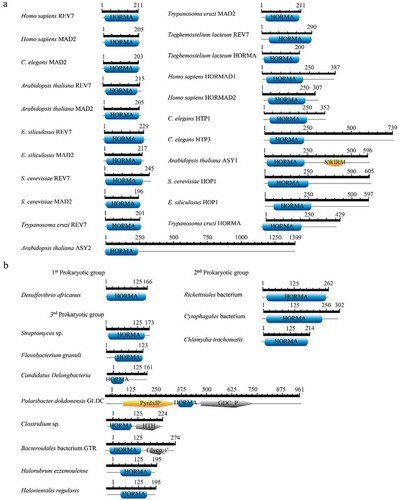
Figure 3. Multiple sequence alignments of HORMA domains from 29 eukaryotic, 20 bacterial and two archaeal HORMA-containing proteins. The proteins are designated by the genus name followed by the name of protein. Uncharacterized proteins are named by genus name followed by “HORMA”. Streptomyces purpurogeneiscleroticus # A0A0M8ZCY4 named as “StreptomycesHORMA”, Streptomyces sp. # A0A2A2Z569 named as “StreptomycesHORMA1”. Alignment was performed using MUSCLE 3.8 and visualized by UniGene.

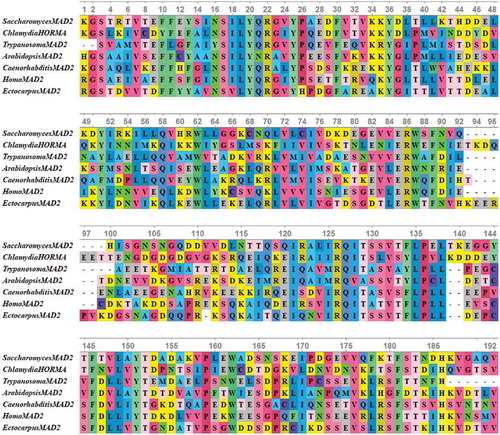
Figure 4. Multiple sequence alignments of conserved HORMA domain in some eukaryotic MAD2 protein sequences along with HORMA domain from Chlamydia trachomatis protein. The proteins are designated by the genus name followed by the name of protein. Alignment was performed using MUSCLE 3.8 and visualized by UniGene.