Figures & data
Figure 1. hNOC3 interacts with hORC and hMCM subunits in yeast two-hybrid assays. The AH109 host yeast cells were co-transformed with the indicated combinations of plasmids, and the transformants were patched onto SCM-Trp-Leu (SCM-2) and SCM-Trp-Leu-His (SCM-3) plates and incubated to test cell growth. Cells containing AD-T (SV40 large T-antigen) and BD-p53 were used as the positive interaction control. Cells containing the two empty AD and BD vectors, AD-hNOC3+ BD vector, and AD vector+BD-hNOC3 were patched on the top three spots of the plates (labelled as Negative controls). Other negative controls also included individual AD-ORC/MCM or BD-ORC/MCM plasmids together with the opposite empty vectors.
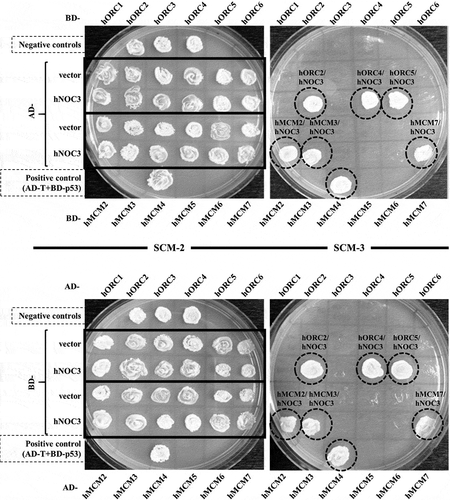
Figure 2. hNOC3 interacts with hMCM subunits in co-IP. HEK-293T cells were transfected with HA-hNOC3 plasmid and harvested for immunoprecipitation after 48 hrs post-transfection. (a) DNase I-treated whole-cell extracts (WCE) were immunoprecipitated with an anti-HA antibody or the control mouse IgG. Co-IP assay results show that hNOC3 interacts with hMCM2, hMCM3 and hMCM7, but not hMCM5. *: Cross-reactive band. (b) Reciprocal co-IP was done using hMCM3 antibody and hNOC3 was co-immunoprecipitated.
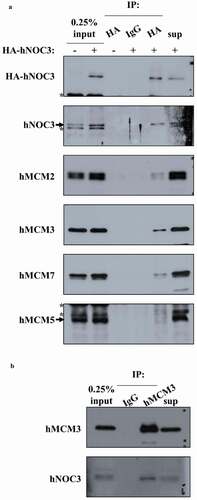
Figure 3. hNOC3 binds to chromatin, preferentially at replication origins, throughout the cell cycle. (a) HeLa cells were synchronized in M-phase by nocodazole followed by mitotic shake-off to obtain mitotic cells, which were then released onto the cell cycle. Cells were then collected at different time points as indicated and analyzed by flow cytometry and chromatin-binding assay to examine the cell cycle progression and chromatin association of hNOC3 and other pre-RC proteins. (b) HEK-293T cells were transfected with HA-hNOC3 plasmid and harvested for ChIP assay after 48 hrs post-transfection. DNA from the immunoprecipitate was analyzed by qPCR using primer sets to quantify the replication origin and non-origin control sequences. α-HA, IP by α-HA antibody; IgG, mock IP by non-specific mouse IgG; UT, IP from untagged hNOC3 control extracts. (**: p < 0.01; ***: p < 0.001.
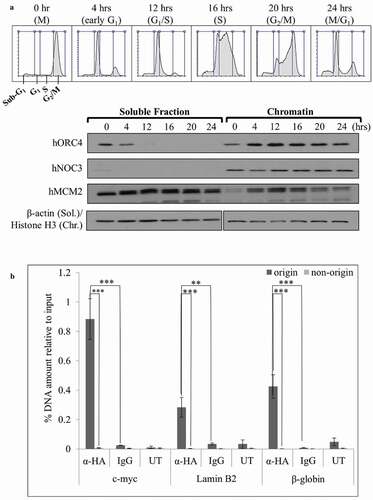
Figure 4. Pre-RC assembly is abrogated by hNOC3 silencing but not by specific inhibition of the ribosome biogenesis pathway. (a) Pre-RC assembly was abrogated after depletion of hNOC3. HeLa cells were transfected with hNOC3 siRNAs, synchronized in G1-phase by mimosine, and then harvested for chromatin-binding assay. The result for flow cytometry analysis was included showing most of the cells were synchronized in G1-phase. LF, the transfection agent Lipofectamine RNAiMax; siNC, negative control siRNA. (b) Pre-RC assembly was abrogated after depletion of hNOC3 but not by specific inhibition of the ribosome biogenesis pathway. HeLa cells were treated with siRNA targeting hNOC3 or the RNA Pol I subunit RPA194, or treated with the RNA Pol I inhibitor CX-5461. The cells were then synchronized in G1-phase by mimosine and harvested for chromatin-binding assay.
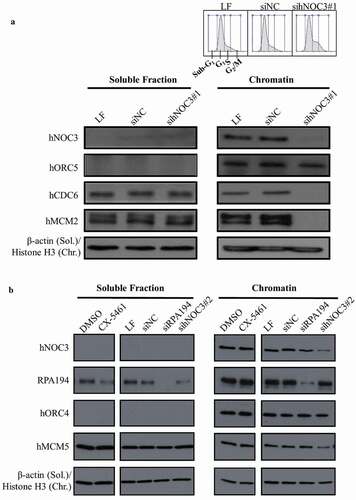
Figure 5. DNA replication is defective after hNOC3 knockdown but not after ribosome biogenesis inhibition. HeLa cells were transfected or treated as indicated and synchronized in G1-phase by mimosine. Cells were released into early S-phase for EdU-incorporation assay. (a) Percentage of EdU-positive cells was reduced after hNOC3 knockdown by either of the two hNOC siRNAs. Flow cytometry result shows that control cells entered S-phase after release while knockdown cells were defective in S-phase entry. LF, Lipofectamine RNAiMax; siNC, negative control siRNA. (b) DNA replication was not affected after specific inhibition of the ribosome biogenesis pathway by siRNA targeting the RNA Pol I subunit RPA194, or by the RNA Pol I inhibitor CX-5461. At least 200 cells/sample were analyzed. (*: p < 0.05).
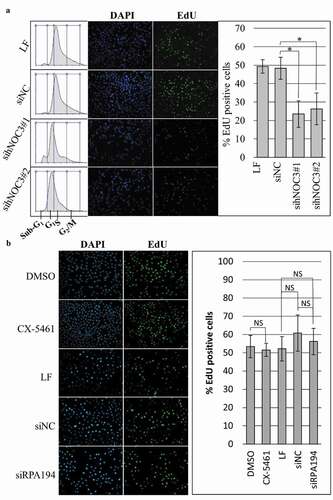
Figure 6. Knockdown of hNOC3 leads to severe apoptosis before S-phase completion HeLa cells were transfected with siRNAs as indicated and synchronized at G1-phase by mimosine. Cells were released and harvested at different time points as indicated. To prevent exit from mitosis, nocodazole was added to a population of the cells at 12 hrs post-release. (a) Results of the Annexin-V apoptosis assay after hNOC3 knockdown. Annexin-Cy3 stains apoptotic cells with disrupted cell membrane, and 6-CFDA stains live cells including early apoptotic cells (early apoptotic cells are positive for both signals). (b) Quantification of the early apoptotic cells in the Annexin-V apoptosis assay in (a). At least 200 cells per sample were examined. (*: p < 0.05; **: p < 0.01; ***: p < 0.001, compared to respective time point of the siNC group) (c) Immunoblotting was done to determine if the cells entered M-phase indicated by the phospho-Histone H3 (serine 10) signal.
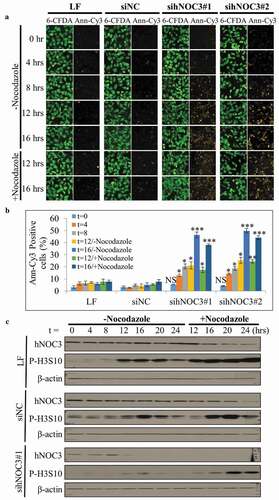
Figure 7. The knockdown of hNOC3, but not specific inhibition of ribosome biogenesis, inhibits S-phase entry and leads to severe apoptosis. HeLa cells were transfected with siRNAs or treated as indicated, and synchronized at G1-phase by mimosine. Cells were released and time points were harvested for flow cytometry analysis. (a) Flow cytometry was performed to examine cell cycle progress and detect apoptotic cells. (b) Flow cytometry result in (a) was confirmed with sihNOC3#2 with a set of simplified time points and compared to that after specific inhibition of the ribosome biogenesis pathway. LF, Lipofectamine RNAiMax; siNC, negative control siRNA.
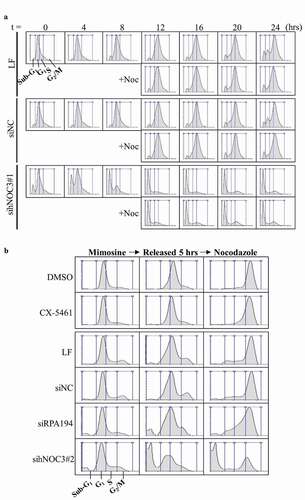
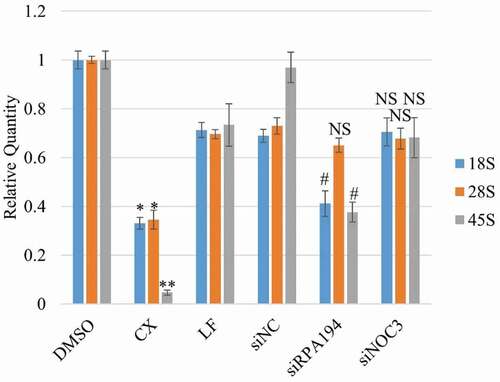
Figure 8. Specific inhibition of Pre-rRNA synthesis results in significant reduction in the 45S precursor rRNA level and the mature 18S and 28S rRNA levels. HeLa cells were treated with siRNAs against hNOC3 or the RNA Pol I subunit RPA194, or treated with the RNA Pol I inhibitor CX5461. Total RNA was isolated and quantified by RT-qPCR using primer sets for 18S, 28S and 45S rRNAs. Cells were harvested at comparable time window as in and . (*: p < 0.05; **: p < 0.01, compared to the DMSO control. #: p < 0.05, compared to the siNC control.).