Figures & data
Table 1. Summary of microarray datasets used in this study
Figure 1. KIF23 expression is significantly up-regulated in ovarian cancer and predicts poor prognosis for ovarian cancer patients.
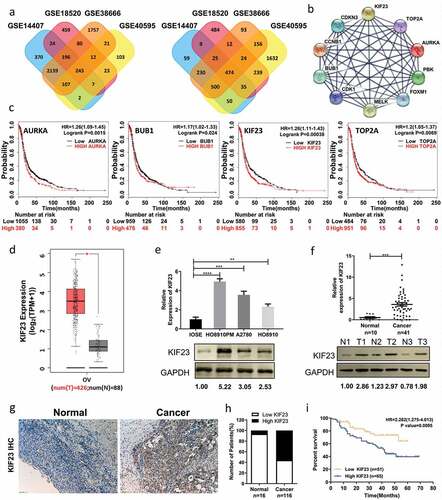
Figure 2. The correlation analyses between KIF23 and cell cycle-related proteins.
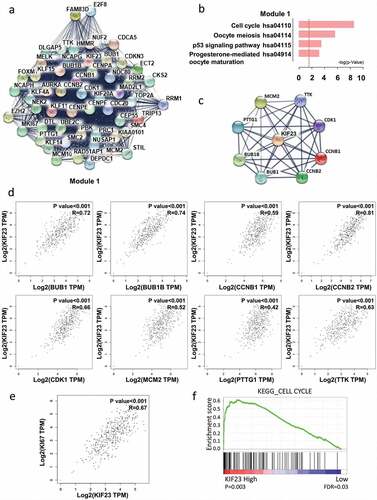
Figure 3. The effect of KIF23 on proliferation, migration and cell cycle.
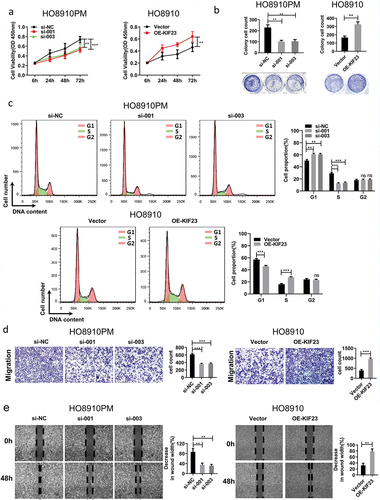
Figure 4. KIF23 is directly targeted by miR-424-5p and miR-503-5p in vitro.
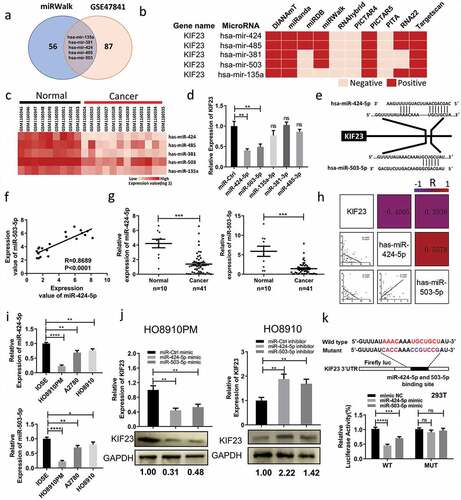
Figure 5. MiR-424-5p and miR-503-5p regulate KIF23 to influence proliferation, migration and cell cycle of ovarian cancer cells.
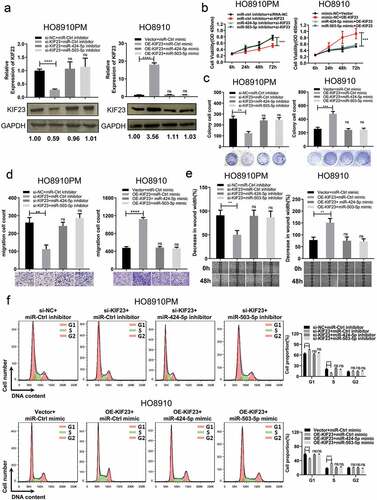
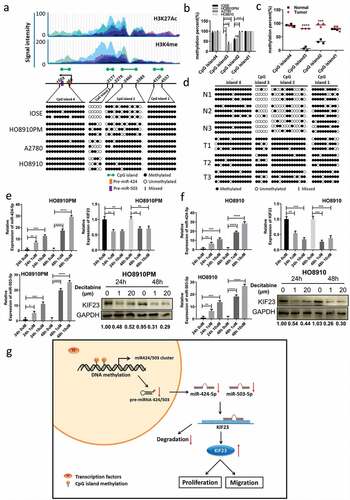
Figure 6. Restoration of miR-424/503 expression and downregulation of KIF23 by demethylation.