Figures & data
Table 1. Genes predicted to be the targets of at least three miR-34a synergistic miRNAs.
Figure 1. Screening approach. (a) Use of a dual luciferase expression vector to evaluate transfection efficiency and cell number. The schematic depicts a model in which firefly luciferase is used as a measurement of the transfection efficiency when cells are co-transfected with a siRNA against firefly luciferase (siLuc2) and a miRNA. Renilla activity serves as a proxy for cell number and its activity is only affected when a miRNA that alters cell proliferation is transfected. (b) A linear response between cell number and Renilla and Firefly luciferase activity (Error bars: mean ± s.d., n = 6). C) Firefly luciferase repression upon transfection with siLuc2. Error bars: mean ± s.d. Each experiment corresponds to n = 6 per treatment. (d) miR-34a dose response in H441-pmiR cells. Error bars: mean ± s.d. Each experiment corresponds to n = 8 per treatment. Statistical analysis performed with two-way ANOVA with post hoc Bonferroni correction (***, P < 0.001). (e) Firefly and (f) Renilla activity following transfection with miR-34a or PremiR-NC2 in presence of siLuc2. Error bars: mean ± s.d. Each experiment corresponds to n = 6 per treatment. Statistical analysis for B, C, D, E and F performed with a one-way ANOVA with post hoc Bonferroni correction (*, P < 0.05; **, P < 0.01; ****, P < 0.0001). RLU: relative light units.
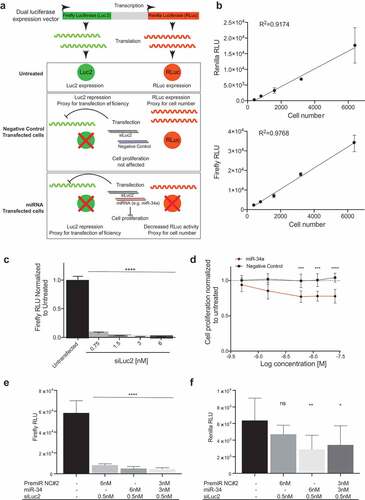
Figure 2. miRNAs alter miR-34a antiproliferative potential. (a) miRNA mimics ranked according to effect, normalized to negative control (NC) transfected cells. Red dashed line = 3 s.d. of the effect of miR-34a alone. Black dashed line = effect of NC treated cells. Blue shaded box = 333 miRNA mimics that were moved forward in B. n = 6. Each dot represents the mean effect of a miRNA combination. (b) Validation of 333 miRNA mimics represented as the effect of the individual miRNA (Esingle) minus the effect when combined with miR-34a (Epair). Values <0 indicate that the combination has a better antiproliferative activity than the miRNA by itself. n = 6. Each dot represents the mean effect of a miRNA combination. Blue shaded box = 111 miRNA mimics with apparent combinatorial activity. The five miRNAs subsequently determined to have synergistic activity with miR-34a are indicated.
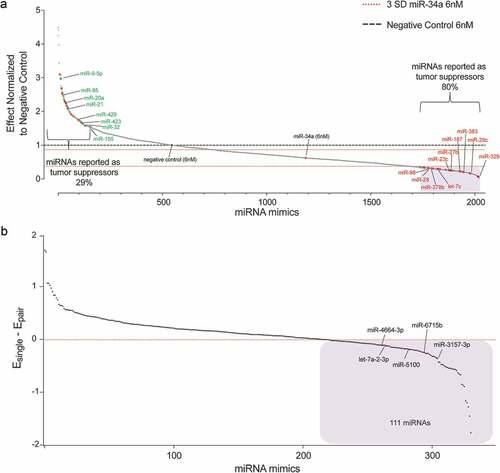
Figure 3. Validation of a combinatorial effect of the 111 miRNA mimics in combination with miR-34a in a panel of NSCLC cell lines. (a) Validation of sulforhodamine B (SRB) method for secondary verification of screen hits. Absorbance of SRB as a linear response to H441-pmiR cell number. Error bars: mean ± s.d., n = 6. (b) Response of H441-pmiR to miR-34a measured by SRB. Error bars: mean ± s.d., n = 6. (c) miRNA combinations evaluated in a panel of six NSCLC cell lines. miRNAs included in this comparison had a CI lower than 1 (potential synergism), p < 0.05 and cell proliferation of the single miRNA was <1.
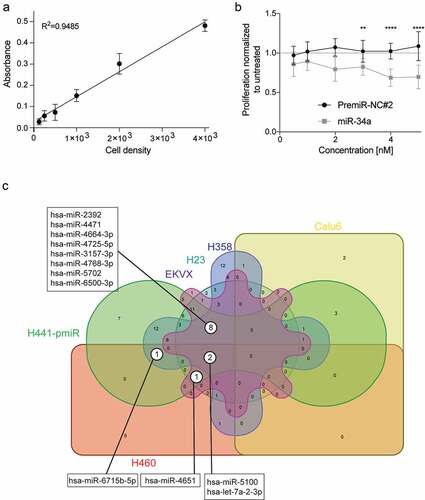
Figure 4. Evaluation of synergism of positive hits. The possibility of synergism with miR-34a was evaluated for (a) miR-6715b, (b) miR-4664-3p, (c) miR-3157-3p, (d) miR-5100 and (e) let-7a-2-3p. (For A-E. red lines: miR-34a alone, blue lines: putative synergistic miRNA alone, green lines: combination) miRNA combinations have a stronger effect than each miRNA on its own. (f) Evaluation of synergism using the (CI)-isobologram equation. The equation helps to gain a better understanding of drug interactions, where CI < 1 indicate synergism. Statistical analysis performed with one-way ANOVA with post hoc Bonferroni correction (***, P < 0.001; ****, P < 0.0001).
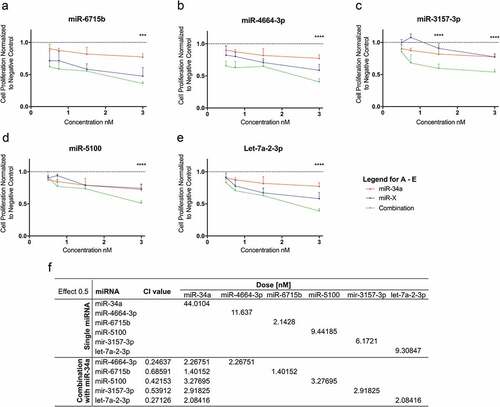
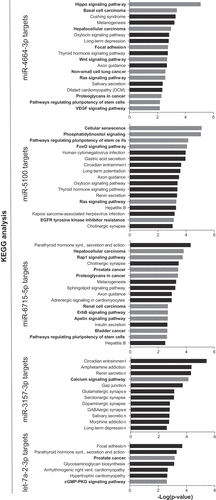
Figure 5. Pathway enrichment analysis of miR-34a-synergistic miRNA target genes. Predicted targets for the five synergistic miRNAs were determined and used to identify pathways that the target genes are enriched in. Cancer-related pathways/diseases are in bold text with corresponding bars depicted in grey.