Figures & data
Figure 1. Proteomic profiling during keratinocyte differentiation. (a) 2-DE proteomic profiling of keratinocytes at 1 d, 3 dand 6 d after calcium treatment. Numbers indicate the statistically variable protein spots as detected by using Progenesis SameSpots software. (b) Venn diagrams depicting the number of proteins identified in each sample, during the calcium treatment. Graphic was obtained by FunRich program submitting the lists of proteins exclusively detected in each sample. (c) Gene Ontology-based functional enrichment analysis of biological functions, using FunRich software.
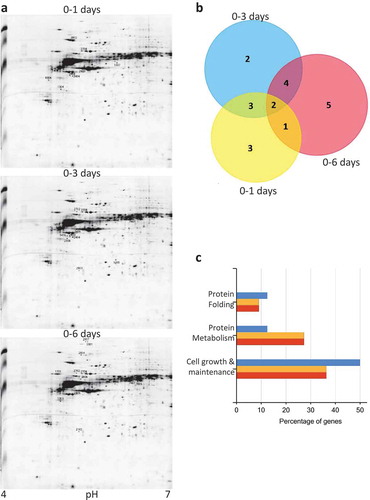
Table 1. List of differentially abundant proteins identified by LC-MS/MS in keratinocytes cells treated by calcium.
Table 2. UPR proteins modulated during keratinocyte differentiation. The table show protein trend and transcriptomic trend.
Figure 2. RAD23 and PDI expression is downregulated during HEKn cells differentiation (a) Western blot of HEKn cells protein extracts collected at calcium-induced keratinocytes differentiation days (DD). Differentiation was evaluated by expression levels of Keratin 10. RAD23 and PDI proteins downregulation during differentiation is also shown. β-Actin was used as loading control. (b) Confocal analysis performed on calcium-induced HEKn differentiation, also reveals a decrease of RAD23 and PDI protein levels (green). Cytoskeleton labeled with β-Tubulin (red) and nuclei stained blue (DAPI).
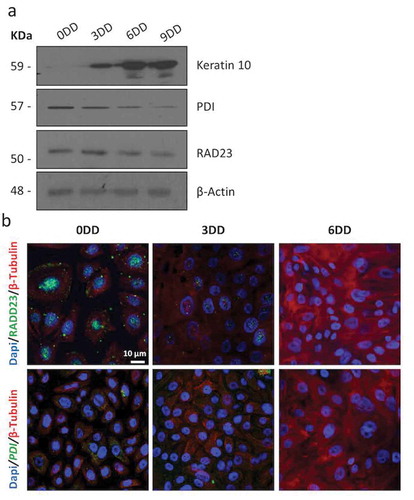
Figure 3. Activation of the UPR during keratinocyte differentiation. (a) Heat map comparing the expression of selected genes using the RNA seq data published in [Citation59] and the ENCODE data. (b) Real-time quantitative PCR showing UPR marker’s, ATF4 AND CHOP, expressed at mRNA level during calcium-induced keratinocytes differentiation. DD: days of differentiation. *p = 0.02; **p = 0.01; ***p = 7x10−7; *p = 0,045; *p = 0,048. (c) Western blot of GRP78 during calcium-induced keratinocytes. DD: days of differentiation. (d) Normal human skin showing, by confocal analysis, GRP78 (red). Basal layer is labeled using K14 (green) and nuclei stained blue (DAPI).
![Figure 3. Activation of the UPR during keratinocyte differentiation. (a) Heat map comparing the expression of selected genes using the RNA seq data published in [Citation59] and the ENCODE data. (b) Real-time quantitative PCR showing UPR marker’s, ATF4 AND CHOP, expressed at mRNA level during calcium-induced keratinocytes differentiation. DD: days of differentiation. *p = 0.02; **p = 0.01; ***p = 7x10−7; *p = 0,045; *p = 0,048. (c) Western blot of GRP78 during calcium-induced keratinocytes. DD: days of differentiation. (d) Normal human skin showing, by confocal analysis, GRP78 (red). Basal layer is labeled using K14 (green) and nuclei stained blue (DAPI).](/cms/asset/c6537eb3-3006-42cf-b703-7049344545fd/kccy_a_1642066_f0003_oc.jpg)
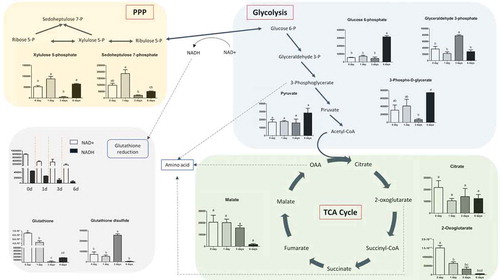
Figure 4. Glycolysis and TCA cycle intermediates during keratinocytes differentiation. Variation in the levels of metabolic intermediates in Glycolysis and Kreb’s cycle. The values shown are mean ± SD (n = 6) of keratinocytes metabolites. Statistical significance was indicated by *p < 0.05; **p < 0.01; ***p < 0.001.
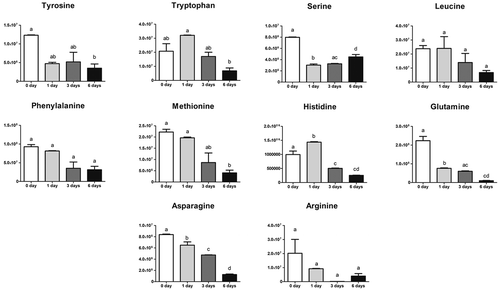
Figure 5. Amino acids during keratinocytes differentiation. Variation in the keratinocytes of amino acids content in keratinocytes. Values are mean ± SD (n = 6) of metabolites. Statistical significance was indicated by *p < 0.05; **p < 0.01; ***p < 0.001.