Figures & data
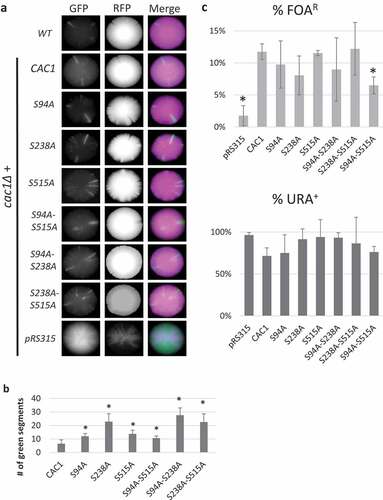
Figure 1. Mutations of the phosphorylation sites of CAC1.
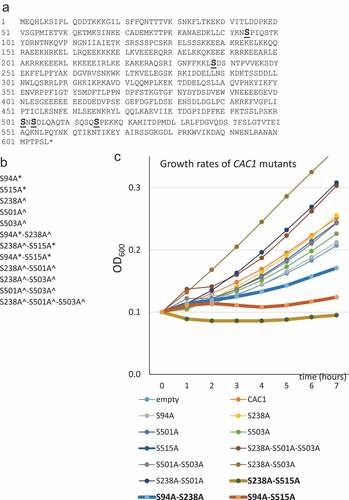
Figure 2. Protein and plasmid stability in CAC1 mutants and association with PCNA
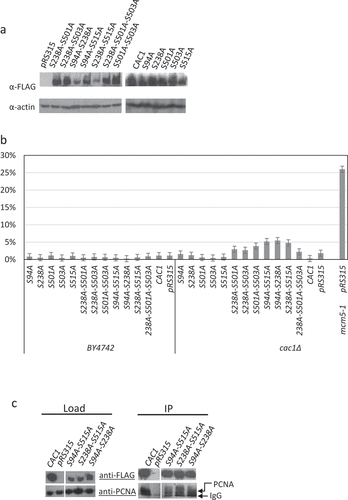
Figure 3. Deregulation of cell cycle in CAC1 mutants
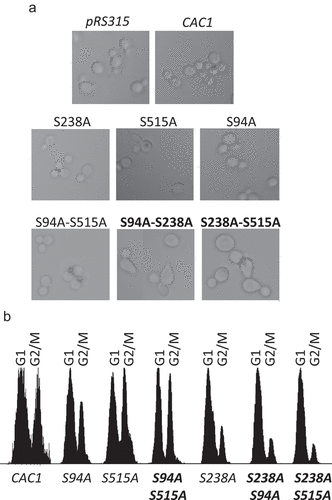
Figure 4. Mutation rates and sensitivity to DNA damage
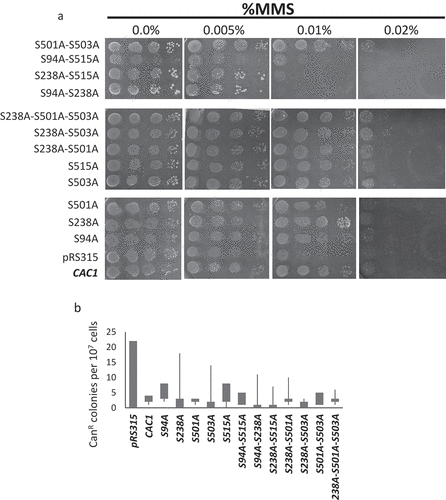
Figure 5. Flocculation in S94A-S238A and S238A-S515A mutants
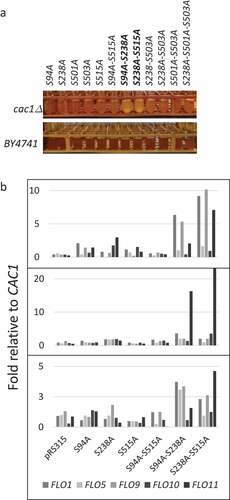
Figure 6. Analysis of silencing at Sir2p-regulated loci