Figures & data
Figure 1. Experiment process. The whole experiment can be divided into four parts: data preprocessing, the identification of editing sites, genetic variants annotation and getting cell-specific editing sites
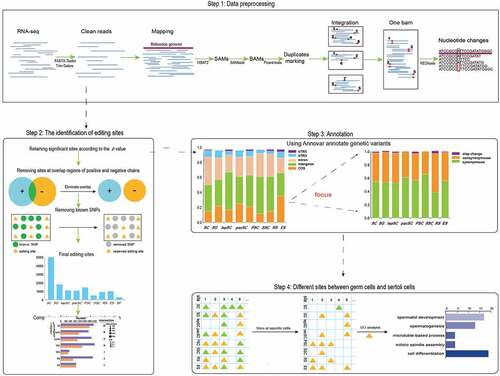
Table 1. The details of the editing sites identified in specific cells
Figure 2. Editing sites identified in different types of cells during spermatogenesis. (a) The number of editing sites for specific types of cells. (b) Distribution of RNA editing levels in different germ cells. (c) Comparison of RNA editing levels between sertoli cells and male germ cells. Statistical significance was determined using t-test, ****p< 0.0001 (d) The relationship between sample size and REs number. (e) Modular RNA editing sites based on cell type. (f) Editing levels of seven genes that only undergo RNA editing in specific germ cells
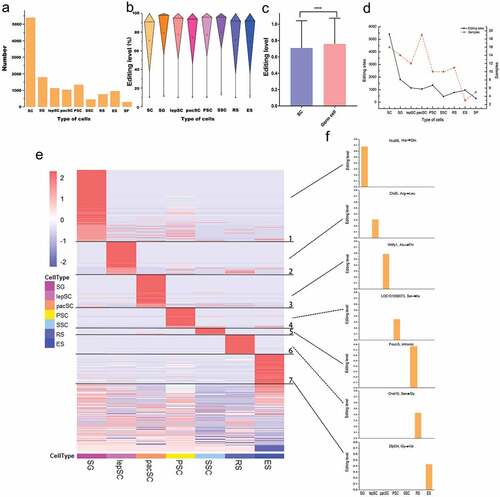
Figure 3. The number of 12 kinds of nucleotide changes. (a) Nucleotide changes in germ cells. (b) Nucleotide changes in sertoli cells. (c) Distribution of editing levels of four main types RNA editing in different germ cells. (d) Comparison of editing levels of these four main RNA editing. Statistical significance was determined using ANOVA, ****p < 0.0001, ns: non-significant
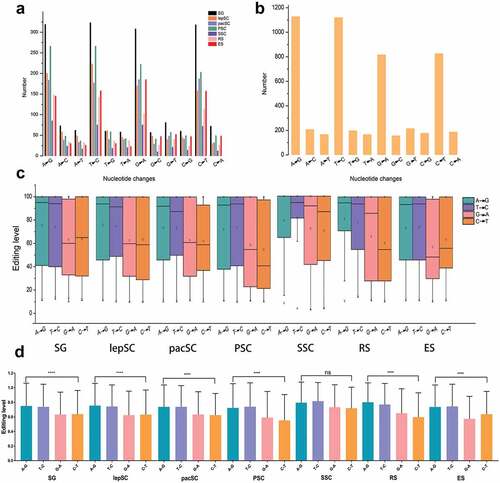
Table 2. Editing sites in genomic repetitive regions
Figure 4. Editing sites on chromosome 17. (a) Distribution of editing sites on each chromosome in all types of cells and RNA editing levels in red bars. Cells are shown in concentric circles and ordered as follow from the inside: SC, SG, lepSC, pacSC, PSC, SSC, RS and ES. (b) The number of editing sites on the unit chromosome length of 21 chromosomes in each type of cell. (c) Function of edited genes on chromosome 17. (d) The expression of Rbmy in different tissues
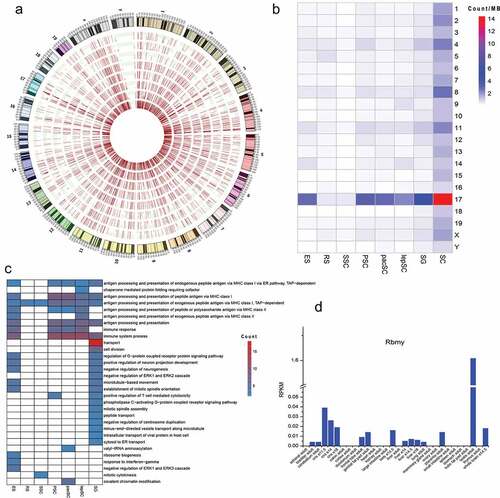
Figure 5. Genes with different numbers of editing events. (a) Proportion of genes with one editing event and genes with two or more editing events in germ cells. (b) Proportion of genes with one editing event and genes with two or more editing events in sertoli cells. (c) Proportion of genes with one nonsynonymous editing event and genes with two or more nonsynonymous editing events in germ cells. (d) The number of genes with different numbers of editing events in germ cells. (e) The number of genes with different numbers of nonsynonymous editing events in germ cells. (f) The expression of Gm5458across different tissues. (g) The expression of Gm21119across different tissues
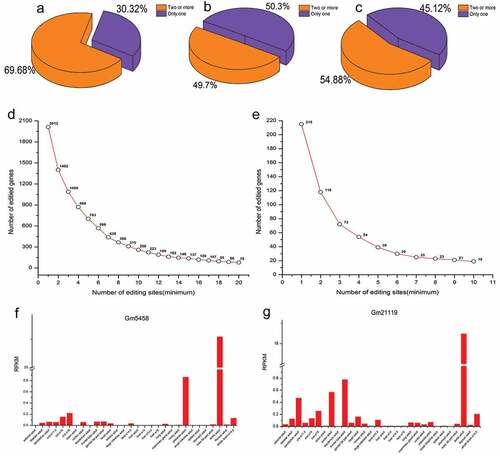
Figure 6. About half of the editing events in CDSs result in amino acid changes. (a) Distribution of the editing sites. (b) Functional consequences of the editing sites. (c) Distribution of the editing sites on chromosome 17
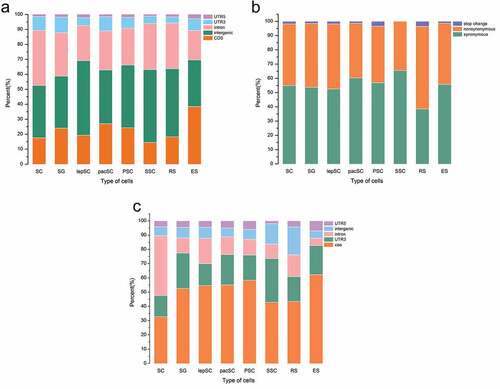
Figure 7. Significant editing events for spermatogenesis. (a) All sites in male germ cells and editing sites only in germ cells. (b) Function of genes that have editing sites only in germ cells. (c) The function of genes which are edited in multiple stages
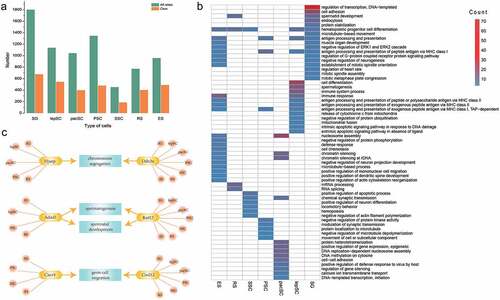
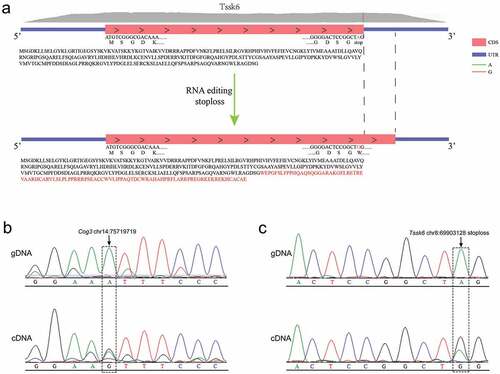
Figure 8. Validation of RNA editing site with Sanger sequencing. (a)The stoploss editing in Tssk6 will change the original stop codon to a codon encoding an amino acid and increase the protein from the original 273 amino acids to 369 amino acids. (b) Sanger sequencing of cDNA and gDNA, at the site chr14:75,719,719 of gene Cog3 the genomic A is highly edited to I. (c) An A-to-I editing occurred at the stop codon of the gene Tssk6.