Figures & data
Figure 1. Structure and function of cyclins and cyclin D1. (a) Schematic representation of the cell cycle. Following mitogenic signals, eukaryotic cells exit quiescence and irreversibly enter in G1 phase after the restriction (R) point. The successive G1, S, G2 and M phases are controlled by cyclins and their cognate CDKs, as indicated. (b) Structure of the CCND1 gene, mRNAs and cyclin D1 proteins. The CCND1 gene (NG-007375.1) comprises five exons (E1-5) separated by four introns (I1-4). It encodes the full-length canonical cyclin D1 product (cyclin D1a) of 295 amino acids (aa). Through alternative splicing, CCND1 generates two types of mRNA: the canonical form (NM_053056.2) and the so-called “b” form, which includes the intron 4 encoding an additional stretch of 33 amino acids. The corresponding protein isoforms, “a” and “b”, are identical over the first 240 amino acids from the N-terminus, but have different C-termini. Isoform “b” lacks the threonine 285 residue and the PEST sequence (aa 241–290) required for degradation, and the LxxLL motif (aa 251–257) required for ligand-dependent interactions with nuclear receptors. By contrast, both isoforms contain the cyclin box required for CDK4/6 and CIP1/KIP1 family binding, and the LxCxE motif (aa 5–9) required for RB1 binding. (c) Schematic representation of the G1-to-S phase transition. For cells to exit quiescence (G0), they require mitogenic signals that activate the RAS signaling pathways and, to a lesser extent, the Wnt/β-catenin and NF-κB pathways. Cyclin D1 is activated at several levels after its translation: stability, assembly with its CDK4/6 partners, and import into the nucleus via the CKIs of the CIP/KIP family. Cyclin D1/CDK4/6 complexes accumulate during the G1 phase, until the start of DNA replication. Cyclin D1 is then exported to the cytoplasm, where it is degraded by UPS. Cyclin D1/CDK4/6 phosphorylates and inactivates RB1 (and pocket proteins RBL1/2, p107 and p130) facilitating the dissociation of E2F transcription factors and the activation of the necessary genes for DNA synthesis. Cyclin D1 degradation is required for the progression of S phase. The GSK3β kinase phosphorylates cyclin D1, which is then taken up by XPO1 and exits the nucleus. Specific E3 ubiquitin ligases ubiquitinylate cyclin D1, which is then degraded by the proteasome machinery 66 [Citation66]
![Figure 1. Structure and function of cyclins and cyclin D1. (a) Schematic representation of the cell cycle. Following mitogenic signals, eukaryotic cells exit quiescence and irreversibly enter in G1 phase after the restriction (R) point. The successive G1, S, G2 and M phases are controlled by cyclins and their cognate CDKs, as indicated. (b) Structure of the CCND1 gene, mRNAs and cyclin D1 proteins. The CCND1 gene (NG-007375.1) comprises five exons (E1-5) separated by four introns (I1-4). It encodes the full-length canonical cyclin D1 product (cyclin D1a) of 295 amino acids (aa). Through alternative splicing, CCND1 generates two types of mRNA: the canonical form (NM_053056.2) and the so-called “b” form, which includes the intron 4 encoding an additional stretch of 33 amino acids. The corresponding protein isoforms, “a” and “b”, are identical over the first 240 amino acids from the N-terminus, but have different C-termini. Isoform “b” lacks the threonine 285 residue and the PEST sequence (aa 241–290) required for degradation, and the LxxLL motif (aa 251–257) required for ligand-dependent interactions with nuclear receptors. By contrast, both isoforms contain the cyclin box required for CDK4/6 and CIP1/KIP1 family binding, and the LxCxE motif (aa 5–9) required for RB1 binding. (c) Schematic representation of the G1-to-S phase transition. For cells to exit quiescence (G0), they require mitogenic signals that activate the RAS signaling pathways and, to a lesser extent, the Wnt/β-catenin and NF-κB pathways. Cyclin D1 is activated at several levels after its translation: stability, assembly with its CDK4/6 partners, and import into the nucleus via the CKIs of the CIP/KIP family. Cyclin D1/CDK4/6 complexes accumulate during the G1 phase, until the start of DNA replication. Cyclin D1 is then exported to the cytoplasm, where it is degraded by UPS. Cyclin D1/CDK4/6 phosphorylates and inactivates RB1 (and pocket proteins RBL1/2, p107 and p130) facilitating the dissociation of E2F transcription factors and the activation of the necessary genes for DNA synthesis. Cyclin D1 degradation is required for the progression of S phase. The GSK3β kinase phosphorylates cyclin D1, which is then taken up by XPO1 and exits the nucleus. Specific E3 ubiquitin ligases ubiquitinylate cyclin D1, which is then degraded by the proteasome machinery 66 [Citation66]](/cms/asset/5d727a98-b31c-4093-89cf-94920d07ac77/kccy_a_1706903_f0001_c.jpg)
Figure 2. Deregulation of CCND1 in human cancers. (a) The frequencies of mutation, amplification, and deletion in human cancers are depicted in the graph. Data were obtained from the cBioPortal for Cancer Genomics (//www.cbioportal.org/). We analyzed 10,259 samples from 9,682 patients with 25 different cancers from the Cancer Genome Atlas. (b) The types and frequencies of missense, in-frame and truncating mutations of CCND1 observed with regards to the cyclin D1 protein structure are shown
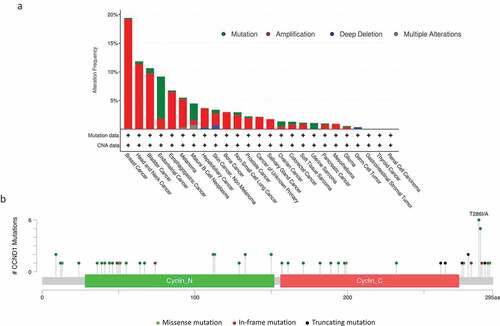
Figure 3. Nuclear functions of cyclin D1. Cyclin D1 activity associated with CDK4/6 is the canonical function of this protein in the control of the cell cycle and cell proliferation. Once activated, cyclin D1/CDK4/6 complexes phosphorylate RB1 (and RBL1/2), leading to their inactivation and the release of E2F family members. The transcriptional program controlled by E2F includes a battery of genes required for G1-to-S phase transition [Citation45]. In addition to its role in cell cycle regulation independent of kinase activity, cyclin D1 acts in complexes with CDK4/6, to sequester the CKIs p21CIP1 and p27KIP1, thereby indirectly controlling the activity of cyclin E/CDK2 complexes acting during the transition G1-to-S. Cyclin D1/CDK4/6 complexes phosphorylate SMAD3 and downregulate the transcription of genes involved in growth inhibition from TGF-β family [Citation68]. Cyclin D1/CDK4 complexes phosphorylate the MEP50 cofactor for PRMT5, an arginine methyltransferase controlling methylation and transcriptional repression. In a mouse model of B-cell lymphomagenesis, Aggarwal and coworkers showed that nuclear cyclin D1 triggered an increase in MEP50/PRMT5 activity, decreasing the activity of CUL4, the E3 ligase of CDT1, the replication licensing protein. Moreover, PRMT5 appears to be necessary for cyclin D1-mediated transformation [Citation119]. However, unlike the targeting of RB1, SMAD3 and FOXM1, this seems to be important during the S phase. We did not comment on this point in the text, but we recommend another recent review including this aspect [Citation78]. Forkhead box M1 (FOXM1) is a critical target of cyclin D1/CDK4/6. Once stabilized by phosphorylation, FOXM1 maintains G1/S phase expression, and protects cancer cells from senescence [Citation69]. FOXM1 appears to be an oncogenic driver for solid tumors [Citation120]. Independently of CDK4/6, cyclin D1 upregulates (ERα) or downregulates (AR, PPARs) nuclear receptor-mediated transcription by binding directly to the receptors and their co-activators (SRC1, AIB1). This regulation may occur directly or through the recruitment of chromatin modifiers of the histone deacetylase or acetyltransferase families (HDAC, HAT) and SuV39H, HP1α. Finally, cyclin D1 binds to proteins of the DNA repair machinery (BRCA1/2, RAD51) and participates to both NHEJ and HR mechanisms of DNA repair
![Figure 3. Nuclear functions of cyclin D1. Cyclin D1 activity associated with CDK4/6 is the canonical function of this protein in the control of the cell cycle and cell proliferation. Once activated, cyclin D1/CDK4/6 complexes phosphorylate RB1 (and RBL1/2), leading to their inactivation and the release of E2F family members. The transcriptional program controlled by E2F includes a battery of genes required for G1-to-S phase transition [Citation45]. In addition to its role in cell cycle regulation independent of kinase activity, cyclin D1 acts in complexes with CDK4/6, to sequester the CKIs p21CIP1 and p27KIP1, thereby indirectly controlling the activity of cyclin E/CDK2 complexes acting during the transition G1-to-S. Cyclin D1/CDK4/6 complexes phosphorylate SMAD3 and downregulate the transcription of genes involved in growth inhibition from TGF-β family [Citation68]. Cyclin D1/CDK4 complexes phosphorylate the MEP50 cofactor for PRMT5, an arginine methyltransferase controlling methylation and transcriptional repression. In a mouse model of B-cell lymphomagenesis, Aggarwal and coworkers showed that nuclear cyclin D1 triggered an increase in MEP50/PRMT5 activity, decreasing the activity of CUL4, the E3 ligase of CDT1, the replication licensing protein. Moreover, PRMT5 appears to be necessary for cyclin D1-mediated transformation [Citation119]. However, unlike the targeting of RB1, SMAD3 and FOXM1, this seems to be important during the S phase. We did not comment on this point in the text, but we recommend another recent review including this aspect [Citation78]. Forkhead box M1 (FOXM1) is a critical target of cyclin D1/CDK4/6. Once stabilized by phosphorylation, FOXM1 maintains G1/S phase expression, and protects cancer cells from senescence [Citation69]. FOXM1 appears to be an oncogenic driver for solid tumors [Citation120]. Independently of CDK4/6, cyclin D1 upregulates (ERα) or downregulates (AR, PPARs) nuclear receptor-mediated transcription by binding directly to the receptors and their co-activators (SRC1, AIB1). This regulation may occur directly or through the recruitment of chromatin modifiers of the histone deacetylase or acetyltransferase families (HDAC, HAT) and SuV39H, HP1α. Finally, cyclin D1 binds to proteins of the DNA repair machinery (BRCA1/2, RAD51) and participates to both NHEJ and HR mechanisms of DNA repair](/cms/asset/0fdbe2b9-2df4-4b2a-8f76-1b645ce1738d/kccy_a_1706903_f0003_c.jpg)
Table 1. Cytoplasmic functions of cyclin D1 in adhesion, migration and invasion
Table 2. Pathway enrichment analysis of cyclin D1 partners in human cancer cell lines
