Figures & data
Figure 1. Dysregulated expression profiles of lncRNAs in NPC. (a) Aberrantly expression of lncRNAs was evaluated in GSE12452. (b, c) The top 10 upregulated and downregulated GO functions of the cellular component (CC) domain. (d, e) The top 10 upregulated and downregulated GO functions of the molecular function (MF) domains. (f) LINC01385 expression was increased in NPC tissues from TCGA dataset. (g, h) High LINC01385 expression was associated with advanced tumor stage and metastasis. (i, j) High LINC01385 expression was associated with poor OS and DFS in NPC patients. *P < 0.05.
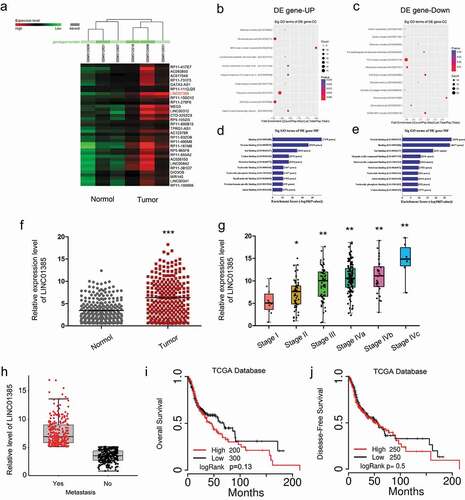
Figure 2. LINC01385 is upregulated in NPC. (a) LINC01385 expression was upregulated in NPC tissues. (b, c) High LINC01385 expression was associated with advanced tumor stage and metastasis. (d) LINC01385 expression was associated with poor OS in NPC patients. (e) LINC01385 was significantly increased in NPC cell lines compared with NP69 cells. (f, g) LINC01385 expression was measured in NPC cells transfected with si-LINC01385 or si-NC by qRT-PCR. *P < 0.05.
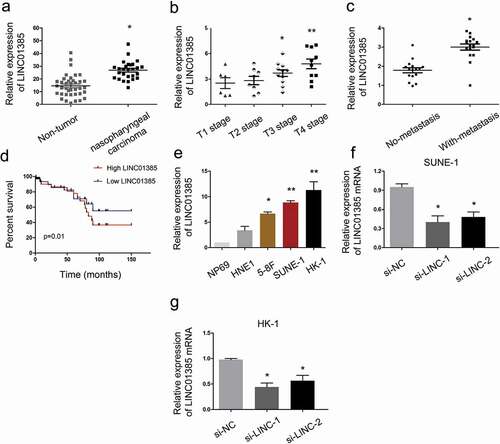
Figure 3. LINC01385 knockdown reduces NPC cells proliferation. (a, b) MTT assay was used to explore the proliferation of SUNE1 and HK-1 cells transfected with si-LINC01385 or si-NC. (c, d) EdU assay was used to determine SUNE1 and HK-1 cells viability in vitro. (e, f) Colony formation assays were explored to test the effects of si-LINC01385 on NPC cells colony formation ability. *P < 0.05.
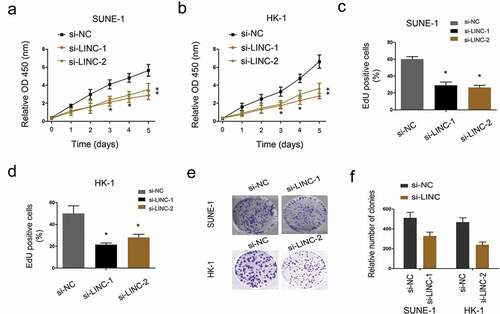
Figure 4. LINC01385 knockdown reduced NPC cells metastasis in vitro. (a, b) Wound healing assay was used to explore the migration ability of SUNE1 and HK-1 cells transfected with si-LINC01385 or si-NC. (c, d) Transwell invasion assay was used to explore the invasion ability of SUNE1 and HK-1 cells transfected with si-LINC01385 or si-NC. (e, f) The effects of si-LINC01385 on Twist1, Snail1 and N-cadherin expression levels were determined by Western blot. *P < 0.05.
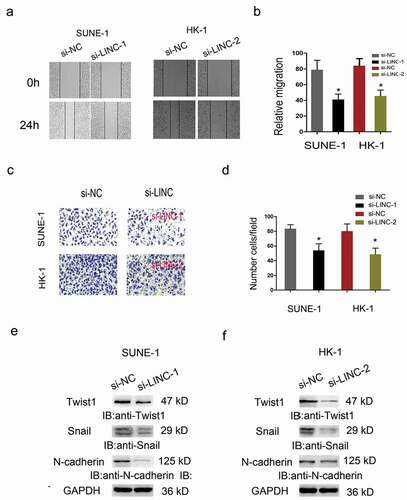
Figure 5. LINC01385 acts as a sponge for miR-140-3p. (a, b) The information about LINC01385. (c) Venn diagrams showed the potential targets of LINC01385. (d) The predicted complementary binding sites between LINC01385 and miR-140-3p. (e-g) Low miR-140-3p expression was significantly associated with advanced tumor stage, metastasis, and poor overall survival rate in NPC patients. (h) Luciferase reporter assay for the luciferase activity in LINC01385-Wt and LINC01385Mut. (i) LINC01385 inhibition induced miR-140-3p expression in NPC cells. (j) LINC01385 expression was negatively correlated with miR-140-3p expression in NPC tissues. *P < 0.05.
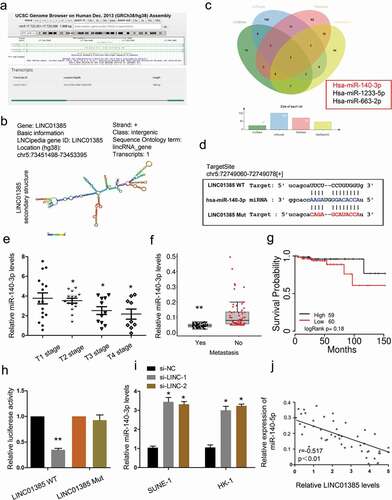
Figure 6. Twist1 is a target gene of miR-140-3p. (a-b) Predicted binding sites for miR-140-3p in the 3ʹUTR of Twist1. (c) The relative luciferase activity was reduced in Twist1-Wt group compared to Twist1-Mut group. (d, e) miR-140-3p mimics significantly reduced Twist1 expression (mRNA and protein levels) in HK1 cells, while Twist1 overexpression abolished the effects. (f) Twist1 expression was upregulated in NPC patients with metastasis by IHC. (g, h) High Twist1 expression was associated with poor OS and DFS in NPC patients. *P < 0.05.
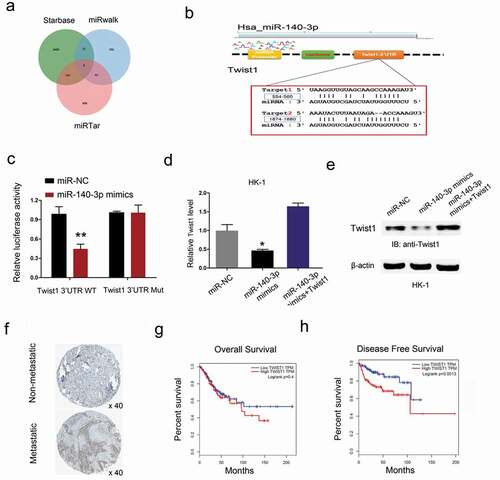
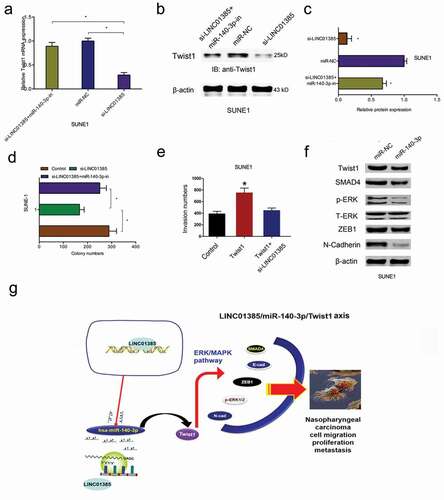
Figure 7. LINC01385 promotes NPC progression through miR-140-3p/Twist1 axis. (a-c) miR-140-3p inhibitors rescued the effects of LINC01385 knockdown on Twist1 expression both in mRNA and protein levels. (d) The anti-colony formation effects of LINC01385 suppression on SUNE1 cells could be abolished by miR-140-3p inhibitors. (e) Twist1 overexpression on SUNE1 cells invasion ability was rescued by LINC01385 suppression. (f) miR-140-3p mimics reduced ERK/MAPK pathway. (g) Schematic diagram of LINC01385 in NPC progression. *P < 0.05.