Figures & data
Figure 1. Decreased expression of GAS5, PDCD4 and PTEN and increased miR-21 level in tissues from MI model. (a) The representative images of TTC staining of sham- or MI-treated rat heart tissues. (b and c) The TUNEL assay of the tissue apoptosis in the rat heart. (d) The images of HE staining and Masson staining of sham- or MI-treated rat heart tissues. (e) ELISA detection of cTnl content in sham- or MI-treated rat serum. (f) The activities analysis of CK, CK-MB and LDH in sham- or MI-treated rat serum. (g) The expression levels of lncRNA-GAS5, miR-21, PDCD4 and PTEN in sham- or MI-treated rat heart tissues. *p < 0.05 and **p < 0.01.
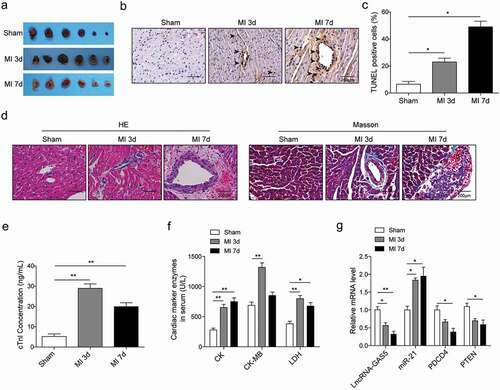
Figure 2. GAS5 regulates H/R-induced cardiomyocyte proliferation and apoptosis. (a) The qPCR analysis of expressions of lncRNA-GAS5, miR-21, PDCD4 and PTEN in H9c2 cells induced by hypoxia for 4–20 h followed by reoxygenation for 3 h. (b) The qPCR analysis of lncRNA-GAS5 expression after overexpression of GAS5 (OE-GAS5) or knockdown of GAS5 (shGAS5). (c) Cell survival assessed by MTT method in H9C2 cells. (d) Cell cycle detection by flow cytometry. (e) Cell apoptosis analysis by flow cytometry under different conditions. (f) Representative images of western blot showing the protein expressions of cleaved caspase-9, BCL-2 and BAX in the cells under different conditions. GAPDH was used as a loading control. Data are presented as the mean ± SD from at least three independent experiments. OE-GAS5: overexpression of GAS5; OE-NC: overexpression of empty plasmid; shGAS5: knockdown of GAS5 with shRNA; shNC: shRNA negative control. *P < 0.05, **P < 0.01 and ***P < 0.001.
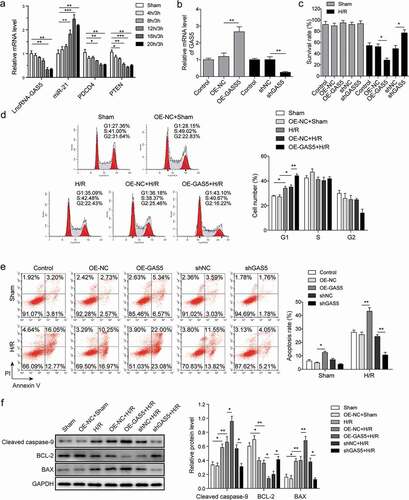
Figure 3. LncRNA GAS5 directly targets miR-21. (a) MiR-21 binding sites in the lncRNA GAS5. Thirteen nucleotides (bold) mutated in luciferase reporter plasmids carrying GAS5. (b) Dual luciferase reporter assay in H9c2 cells transfected with miR-NC or miR-21 mimics followed by the introduction of GAS5-WT or GAS5-MUT. (c) RIP analysis of GAS5 and miR-21 immunoprecipitated by anti-Ago2. The anti-IgG as a negative control. (d) The qPCR analysis of miR-21 level in the cells transfected with OE-GAS5 or shGAS5. Data are presented as the mean ± SD from at least three independent experiments. OE-GAS5: overexpression of GAS5; OE-NC: overexpression of empty plasmid; shGAS5: knockdown of GAS5 with shRNA; shNC: shRNA negative control. *p < 0.05 and **p < 0.01.
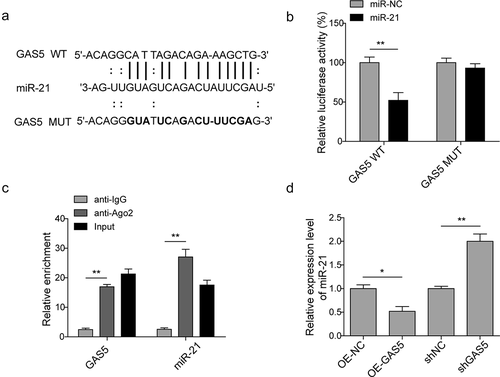
Figure 4. MiR-21 is involved in cardiomyocytes apoptosis induced by lncRNA GAS5. (a) MTT measurement in control or H/R (16 h/3 h)-treated H9c2 cells transfected with miR-21 mimics or GAS5 or co-transfected with miR-NC or miR-21 and GAS5. (b) Cell cycle determination by flow cytometry in the cells treated with OE-NC+H/R, OE-GAS5 + H/R, miR-NC+H/R, miR-21 + H/R, OE-GAS5+ miR-NC+H/R, OE-GAS5+ miR-21 + H/R. (c) Cell apoptosis analysis by flow cytometry in the cells treated with OE-NC+H/R, OE-GAS5 + H/R, miR-NC+H/R, miR-21 + H/R, OE-GAS5+ miR-NC+H/R and OE-GAS5+ miR-21 + H/R. (d) Western blot showing the protein expression of cleaved caspase-9, BCL-2, and BAX in the cells transfected with miR-21 mimics or GAS5 or co-transfected with miR-NC or miR-21 and GAS5 under H/R (16 h/3 h). GAPDH was used as a loading control. Data are presented as the mean±SD from at least three independent experiments. OE-GAS5: overexpression of GAS5; OE-NC: overexpression of empty plasmid; *p < 0.05 and **p < 0.01.
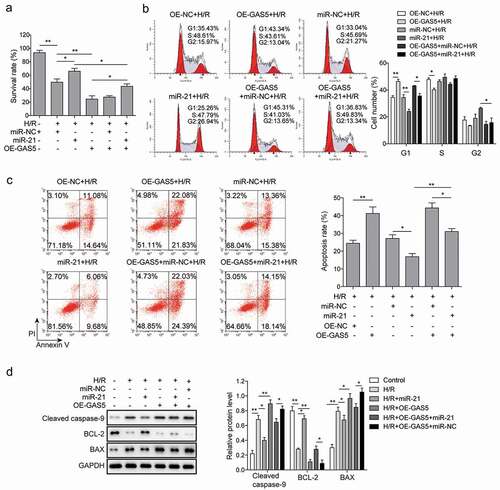
Figure 5. LncRNA GAS5 positively regulates PDCD4 expression by functioning as a sponge of miR-21 in H/R. (a) Dual luciferase reporter assay in the cells transfected with miR-NC or miR-21 and luciferase reporter plasmids carrying 3ʹ-UTR of PDCD4. (b) qPCR analysis of PDCD4 in the cells transfected with GAS5, miR-NC, miR-21 mimics or co-transfected with miR-NC or miR-21 mimics and GAS5 under H/R (16 h/3 h). (c) Western blot showing the protein level of PDCD4 in the cells transfected with GAS5, miR-NC, miR-21 mimics or co-transfected with miR-NC or miR-21 mimics and GAS5 under H/R (16 h/3 h). (d) Western blot was applied to detect the protein level of PDCD4 in the cells transfected with shPDCD4 or GAS5 or co-transfected with shNC or shPDCD4 and GAS5 under H/R (16 h/3 h). (e) Western blot was applied to detect the protein level of PDCD4 in the cells of control, H/R, H/R+ shNC or H/R+ shGAS5 groups. (f) Measurement of cell apoptosis by flow cytometry in the cells transfected with shPDCD4 or GAS5 or co-transfected with shNC or shPDCD4 and GAS5 under H/R (16 h/3 h). (g) Western blot showing the protein expression of cleaved caspase-9, BCL-2, and BAX in the cells under H/R (16 h/3 h). Data are presented as the mean ± SD from at least three independent experiments. *p < 0.05 and **p < 0.01.
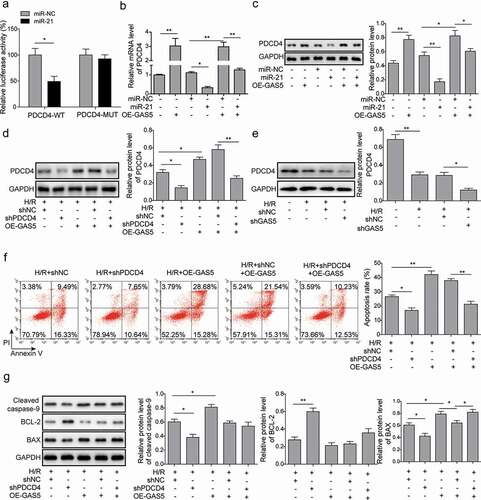
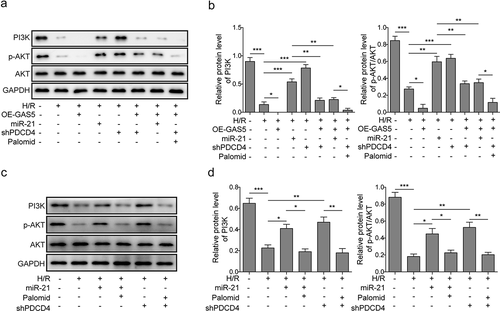
Figure 6. LncRNAGAS5 positively regulates PDCD4 and suppresses PI3 K/AKT signal pathway. (a) Representative image of western blot presenting the protein expression of PI3 K, p-AKT, and AKT in control or H/R(16 h/3 h)-treated cells induced by miR-21 mimics, OE-GAS5, or shPDCD4 or co-transfected by OE-GAS5 and miR-21 or shPDCD4 without or with the AKT inhibitor (Palomid, 2 μM). (b) Densitometry performed for quantification of western blot. GAPDH was used as a loading control. (c) Representative image of western blot presenting the protein expression of PI3 K, p-AKT, and AKT in control or H/R-treated cells or H/R-treated cells induced by miR-21 mimics or shPDCD4 without or with the AKT inhibitor (Palomid, 2 μM). (d) Densitometry performed for quantification of western blot. GAPDH was used as a loading control. Data are presented as the mean ± SD from at least three independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001.
Data availability statement
All data generated or analyzed during this study are included in this published article.
