Figures & data
Figure 2. SLC16A1-AS1 expression and its clinical significance.
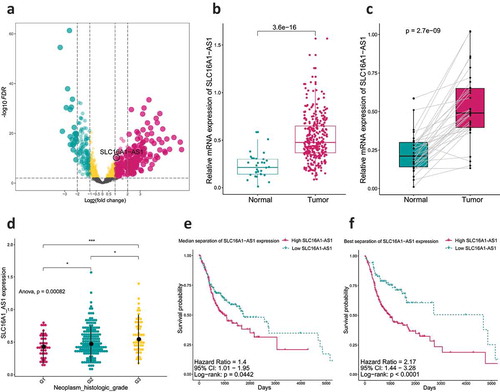
Figure 3. The relationship between SLC16A1-AS1 and tumorigenesis-related features.
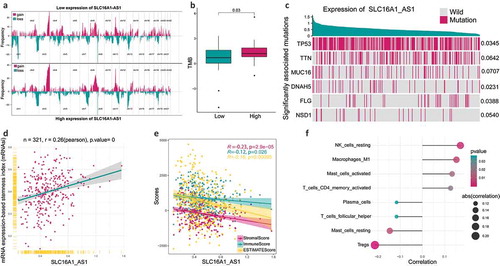
Figure 4. WGCNA identifying the key regulating module and biological functions SLC16A1-AS1 involved in.
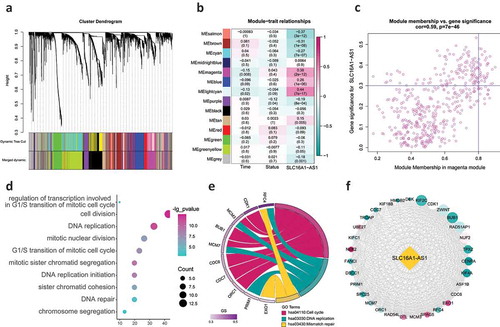
Figure 5. SLC16A1-AS1 is involved in regulation of OSCC cell proliferation via GSEA and GSVA analysis.
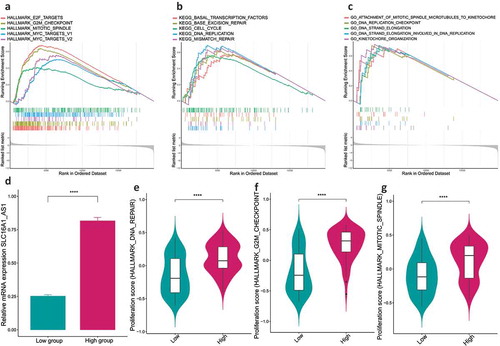
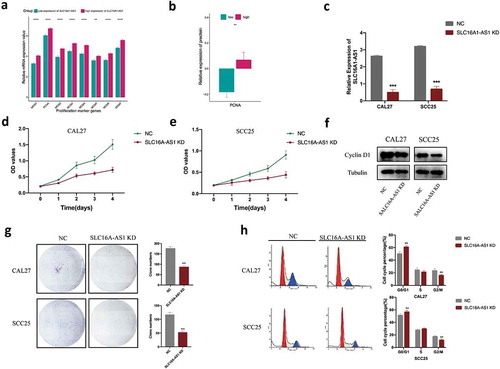
Figure 6. SLC16A1-AS1 silencing inhibits cell proliferation and arrest cell cycle of OSCC cells in vitro.