Figures & data
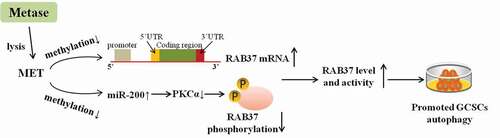
Figure 1. Autophagy was elevated in gastric cancer stem cells (GCSCs)
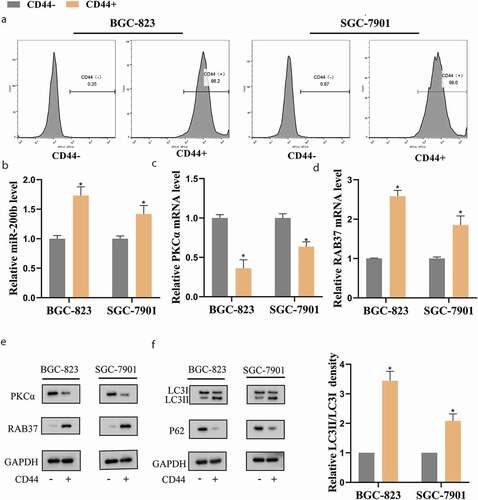
Figure 2. MET increased the methylation and phosphorylation of RAB37 and suppressed autophagy in GCSCs
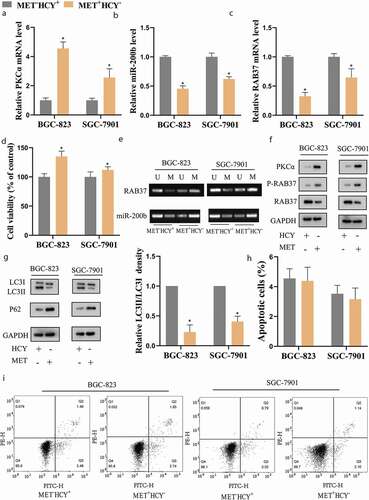
Figure 3. Metase supplementation reduced the methylation and phosphorylation of RAB37 and promoted autophagy in GCSCs
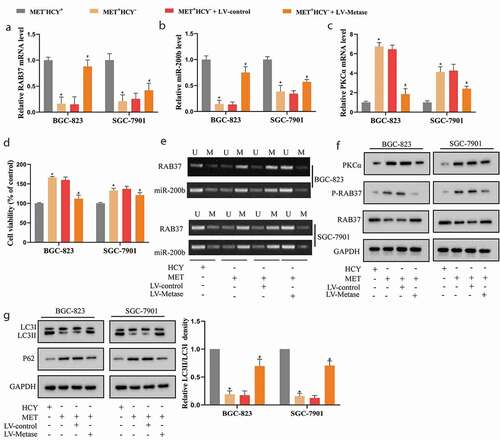
Figure 4. miR-200b negatively regulated PKCα expression in GCSCs
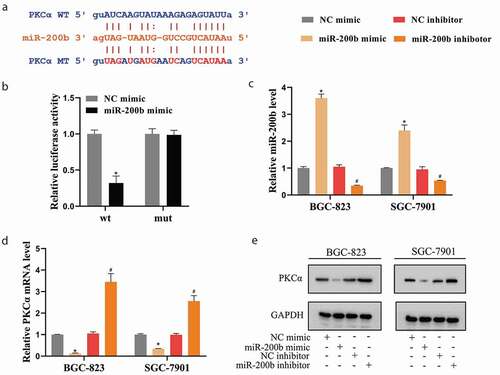
Figure 5. miR-200b/PKCα axis was involved in the promoting effect of Metase on autophagy of GCSCs by regulating the phosphorylation level of RAB37
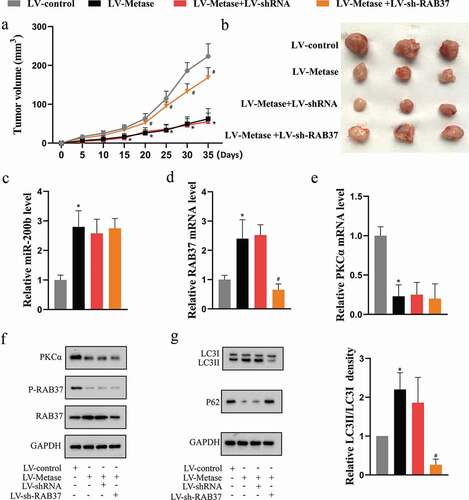
Figure 6. Metase suppressed tumor growth by enhancing the expression and activity of RAB37 in vivo.