Figures & data
Figure 1. CircABCC4 level was more high in breast tumor tissues than in normal breast tissues. The level of circABCC4 was discovered by qRT-PCR. qRT-PCR, Quantitative real-time polymerase chain reaction; (n = 25). ANOVA remained responsible for P-values, ***P < 0.001 was considered as significant results
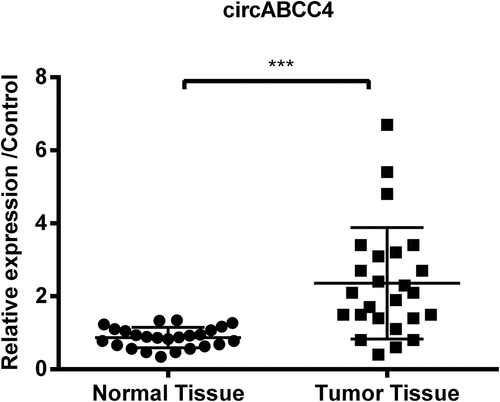
Figure 2. CircABCC4 knockdown repressed breast cancer cells viability and encouraged apoptosis. (a) qRT-PCR analysis for detection of circABCC4 after breast cancer cell lines were transfected with si-NC, si-circABCC4 or corresponding controls. (b) The cell viability was detected by CCK-8. (c) The apoptosis of breast cancer cell lines were identified by flow cytometry. (df) The expression of CyclinD1 as well as CDK6 was investigated by western blot. (gi) The expression of cells apoptosis-correlated proteins (Bcl-2, Bax, Cleaved/Caspase-3) were examined by western blot. CTRL, control; si, small interfering; NC, Negative control; CCK-8, Cell Counting Kit-8; qRT-PCR, Quantitative real-time polymerase chain reaction (n = 25). ANOVA remained responsible for P-values, ** P < 0.01 and *** P < 0.001 were considered as significant results
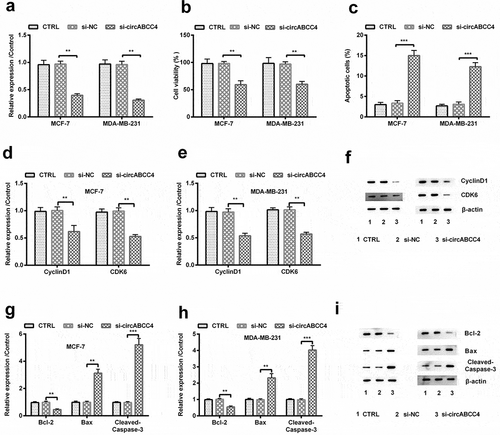
Figure 3. CircABCC4 knockdown inhibited breast cancer cells migration as well as invasion. (ab) The cell migration as well as cell invasion of breast cancer cell lines were detected by Transwell chamber assay. CTRL, control; si, small interfering; NC, Negative control (n = 25). ANOVA remained responsible for P-values, ** P < 0.01 was considered as significant results
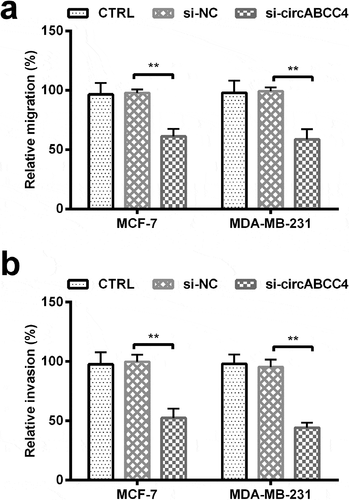
Figure 4. CircABCC4 depressingly controlled the level of miR-154-5p. (a) The complementary binding sites were explored via bioinformatics analysis, and (b) level of miR-154-5p in breast cancer cell lines was measured by RT-qPCR after transfection of si-NC, si-circABCC4 or the corresponding control. TRL, control; si, small interfering; NC, Negative control; miR, microRNA (n = 25). ANOVA remained responsible for P-values, ** P < 0.01 was considered as significant results
Figure 5. CircABCC4 knockdown subdued cell viability and promoted apoptosis of breast cancer cells by promoting miR-154-5p. (a) NC inhibitor and inhibitory miR-154-5p were transfected into breast cancer cell lines as well as the level of miR-154-5p was detected by qRT-PCR. (b) The cell viability was detected by CCK-8. (c) The apoptosis of breast cancer cell lines were detected by flow cytometry. (df) The level of CyclinD1 also CDK6 was analyzed through western blot. (gi) The expression of breast cancer cell lines apoptosis-correlated proteins (Bcl-2, Bax, Cleaved/Caspase-3) were examined through western blot. CTRL, control; si, small interfering; NC, Negative control; miR, microRNA; CCK-8, Cell Counting Kit-8; qRT-PCR, Quantitative real-time polymerase chain reaction; CDK, cyclin-dependent kinases (n = 25). ANOVA remained responsible for P-values, * P < 0.05, ** P < 0.01 and *** P < 0.001 were considered as significant results
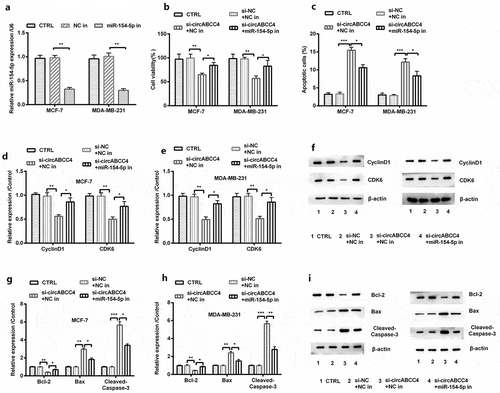
Figure 6. CircABCC4 knockdown inhibited cell migration besides invasion of breast cancer cells by promoting miR-154-5p. (ab) The cell migration also cell invasion of breast cancer cell lines were detected by Transwell chamber assay. CTRL, control; si, small interfering; NC, Negative control; miR, microRNA (n = 25). ANOVA remained responsible for P-values, * P < 0.05 and ** P < 0.01 were considered as significant results
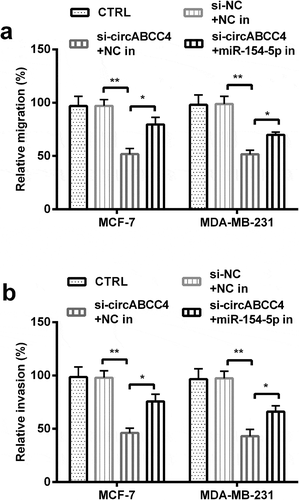
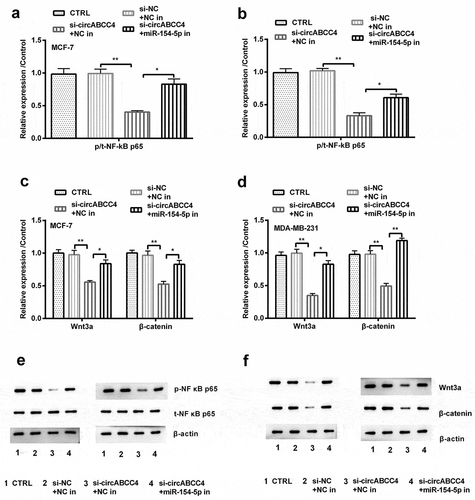
Figure 7. CircABCC4 knockdown inhibited NF-κB and Wnt/β-catenin signal pathway via raising miR-154-5p. (ab) After transfection with si-circABCC4, miR-154-5p inhibitor or their corresponding controls, the protein of the NF-κB and Wnt/β-catenin signal pathways were analyzed by Western blot. CTRL, control; si, small interfering; NC, Negative control; miR, microRNA (n = 25). ANOVA remained responsible for P-values, * P < 0.05 and ** P < 0.01 were considered as significant results