Figures & data
Table 1. Primers used in real-time PCR analysis
Table 2. Weight data of rats
Figure 1. Effect of Gypenosides on liver tissue damage and biochemical indicators in NAFLD rat model. (a) Pathological changes of liver tissue were observed by hematoxylin and eosin (HE) staining in NAFLD rat model that treated with Gyp. (b) The scores were counted according to HE staining. (c) Serum total cholesterol level was detected by serum total cholesterol kit. (d) Triglyceride level was measured by triglyceride kit. (e) Aminotransferase (AST) level was detected by AST kit. (f) Alanine transaminase (ALT) level was detected by ALT kit. (g) Superoxide dismutase (SOD) level was detected by SOD kit. (h) Malondialdehyde (MDA) level was detected by MDA kit. n = 3, *P < 0.05, **P < 0.01, ***P < 0.001, vs. Control; ^P < 0.05, ^^P < 0.01, ^^^P < 0.001, vs. Model; #P < 0.05, ##P < 0.01, ###P < 0.001, vs. Control
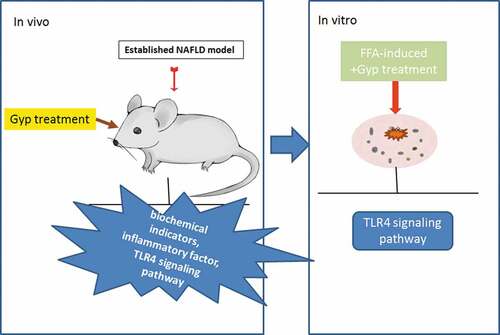
Figure 2. Effect of Gypenosides on biochemical indicators and inflammatory factor and LPS/TLR4 downstream pathway in NAFLD rat model (a) hepatic steatosis index (HSI) of rats was evaluated. (b) Fasting blood glucose (FBG) of rats was measured. (c) Fasting insulin (FINS) of rats was detected. (d) Insulin resistance (HOMA-IR) of rats was evaluated. (e) The expressions of IL-1β, TNF-α, and TLR4 were detected by qRT-PCR. (f) Serum LPS concentration was detected by ELISA kits. (G, H, I and J) The expressions of TLR4, MyD88, p-IκBα, κBα and p-p65 and p65 were detected by Western blotting. (k) QRT-PCR was used to detect the iNOS expression in liver tissue. n = 3, *P < 0.05, **P < 0.01, ***P < 0.001, vs. Control; ^P < 0.05, ^^P < 0.01, ^^^P < 0.001, vs. Model; #P < 0.05, ##P < 0.01, ###P < 0.001, vs. Control
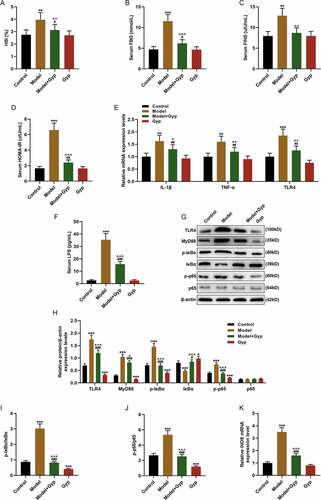
Figure 3. The effect of Gypenosides on FFA-induced adipogenesis in THLE-2 cells and the effect on LPS/TLR4 downstream pathways in THLE-2 cells (a) MTT was used to detect the viability of THLE-2 cell treated with different concentrations of Gyp (10 μmol/L, 20 μmol/L, 40 μmol/L, 80 μmol/L, 160 μmol/L, and 320 μmol/L,). (b and c) Oil Red O staining was used to detect lipid droplet deposition of FFA-induced THLE-2 cell that treated with 40 μmol/L Gyp. (d) Kit used to detect triglyceride (TG) content in cells. (e) The iNOS expression was detected by qRT-PCR in cells. (F, G, H and I) The expression levels of TLR4, MyD88, p-IκBα, κBα, and p-p65 and p65 were detected by Western blotting. (J, K, and L) The p65 expression in cytoplasm and nucleus were detected by Western blotting. n = 3, +P < 0.05, ++P < 0.01, +++P < 0.001, vs. 0; *P < 0.05, **P < 0.01, ***P < 0.001, vs. Control; ^P < 0.05, ^^P < 0.01, ^^^P < 0.001, vs. FFA; #P < 0.05, ##P < 0.01, ###P < 0.001, vs. Gyp; &P < 0.05, &&P < 0.01, &&&P < 0.001, vs. FFA+Gyp+NC
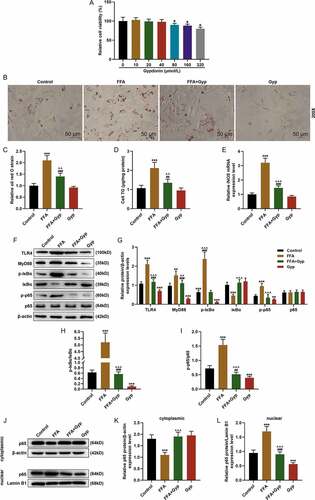
Figure 4. Effects of LPS stimulation or upregulating TLR4 signaling pathway on Gyp’s inhibition of FFA-induced simple steatosis in THLE-2 cells. (a and b) Oil Red O staining was used to detect lipid droplet deposition. (c) Kit used to detect triglyceride (TG) content in cells. (D, E, F, and G) The expression levels of MyD88, p-IκBα, κBα, and p-p65 and p65 were detected by Western blotting. n = 3, +P < 0.05, ++P < 0.01, +++P < 0.001, vs. 0; *P < 0.05, **P < 0.01, ***P < 0.001, vs. Control; ^P < 0.05, ^^P < 0.01, ^^^P < 0.001, vs. FFA; #P < 0.05, ##P < 0.01, ###P < 0.001, vs. Gyp; &P < 0.05, &&P < 0.01, &&&P < 0.001, vs. FFA+Gyp+NC
Data availability statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
