Figures & data
Figure 1. Chromosome clustering in RepoMan-depleted cells during prometaphase
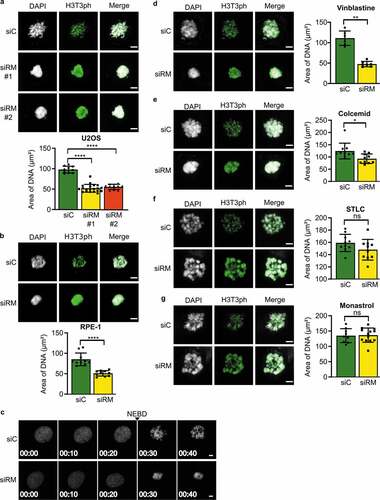
Figure 2. Chromosome clustering in RepoMan-depleted cells is reversible
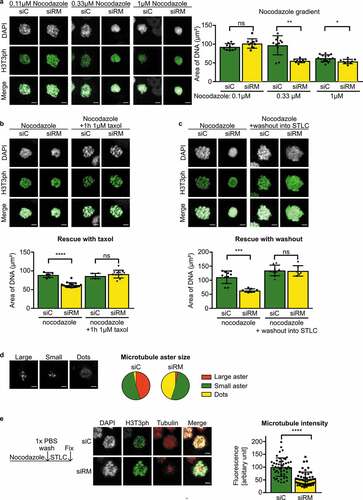
Figure 3. RepoMan interacts with essential microtubule-associated proteins
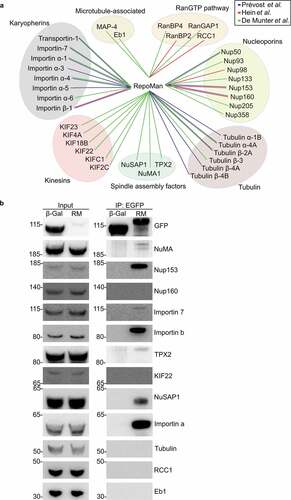
Figure 4. The depletion of RepoMan increases the NuSAP1-importin interaction in a phosphorylation-dependent manner
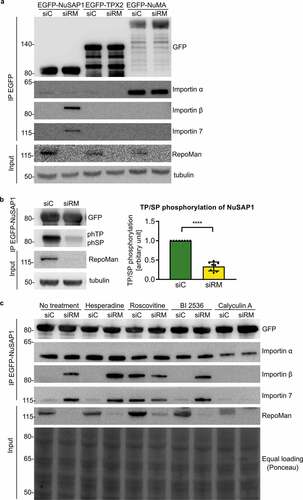
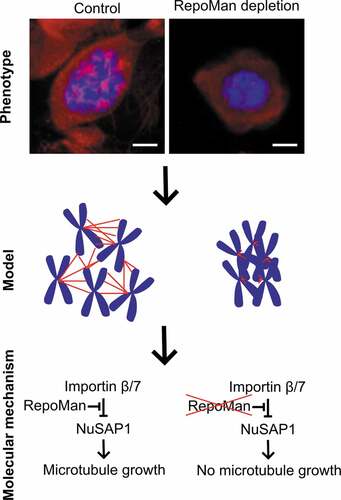
Figure 5. Model on the role of RepoMan in chromosome-dependent spindle assembly
Supplemental Material
Download Zip (4.2 MB)Data Availability Statement
No datasets were generated during the current study.
