Figures & data
Table 1. Gender, stage, and formation reason of keloid patients (18–55 years old)
Figure 1. Circ_0008450 was highly expressed in keratinized epithelial tissues. Keratinized epithelial tissues and normal epithelial tissues were obtained for qRT-PCR experiment. The expression of circ_0008450 was assessed. *** p < 0.001
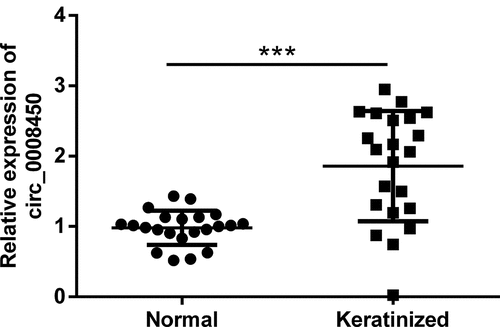
Figure 2. Circ_0008450 silence inhibited the growth of keratinized epithelial cell. Keratinized epithelial cells transfected with sh-circ_0008450 (sh-circ_0008450) or sh-NC were applied for the detection of cell proliferation and apoptosis. (a) The expression of circ_0008450 was assessed by qRT-PCR. *** p < 0.001. Proliferation marker (b) viability and (c) BrdU+ cells were assessed by CCK-8 assay and BrdU incorporation labeling respectively. *** p < 0.001. Apoptosis marker (d) apoptotic rate and (e-f) CyclinD1, CDK4, cleaved caspase 3, and cleaved caspase 9 proteins were assessed by apoptosis assay and western blot respectively. *** p < 0.001
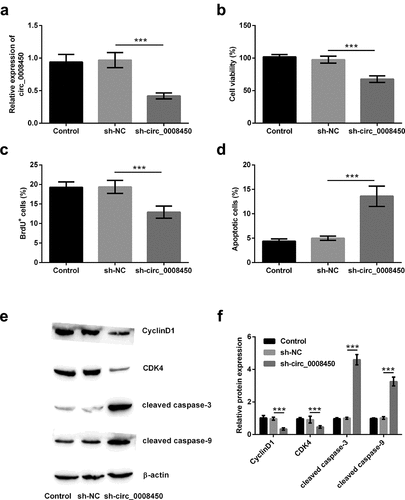
Figure 3. Circ_0008450 silence prevented the migration of keratinized epithelial cell and EMT process. Keratinized epithelial cells transfected with sh-circ_0008450 (sh-circ_0008450) or sh-NC were applied for the detection of migration and EMT process. (a) Relative migratory rate was assessed by migration assay. ** p < 0.01 or *** p < 0.001. (b-c) EMT marker E-cadherin, Vimentin, Fibronectin and ZO-1 were assessed by western blot. ** p < 0.01 or *** p < 0.001
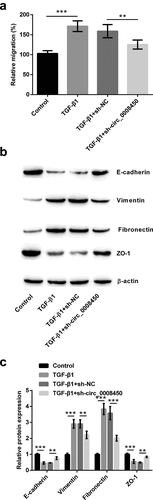
Figure 4. Runx3 was negatively regulated by circ_0008450. Keratinized epithelial cells transfected with sh-circ_0008450 (sh-circ_0008450) or sh-NC were applied for the detection of Runx3 (a) mRNA and (b and c) protein by qRT-PCR and Western blot respectively. *** p < 0.001. (d) Keratinized epithelial tissues and normal epithelial tissues were obtained for qRT-PCR experiment. The expression of Runx3 was assessed by qRT-PCR. *** p < 0.001. (e) Keratinized epithelial tissues were obtained for qRT-PCR experiment. The expression of circ_0008450 and Runx3 was assessed by qRT-PCR. R2 = 0.6909, p < 0.001
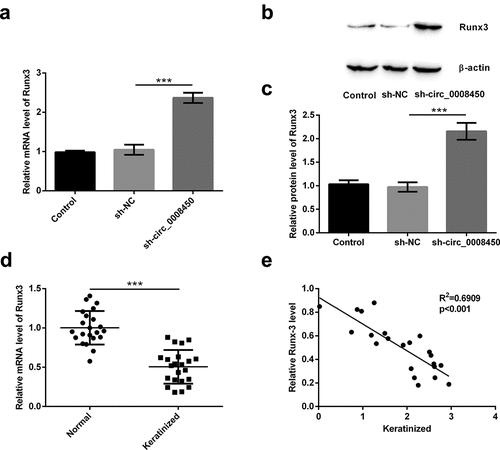
Figure 5. Circ_0008450 silence inhibited the growth of keratinized epithelial cell via the up-regulation of Runx3. (a) Keratinized epithelial cells transfected with sh-Runx3 (sh-Runx3) or sh-control were applied for the detection of Runx3 mRNA by qRT-PCR. *** p < 0.001. Keratinized epithelial cells transfected with sh-circ_0008450 (sh-circ_0008450) or sh-Runx3 (sh-Runx3) or both were applied for the detection of cell proliferation and apoptosis. Proliferation marker (b) viability and (c) BrdU+ cells were assessed by CCK-8 assay and BrdU incorporation labeling respectively. ** p < 0.01 or *** p < 0.001. Apoptosis marker (d) apoptotic rate and (e and f) CyclinD1, CDK4, cleaved caspase 3 and cleaved caspase 9 proteins were assessed by apoptosis assay and western blot respectively. * p < 0.05, ** p < 0.01 or *** p < 0.001
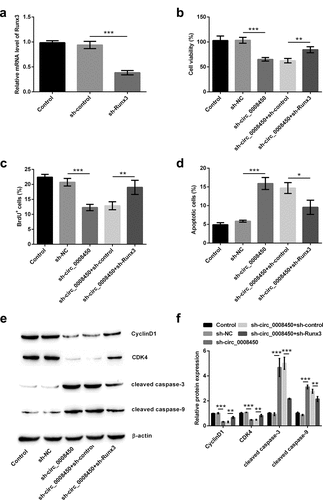
Figure 6. Circ_0008450 silence prevented the migration of keratinized epithelial cell and EMT process via the up-regulation of Runx3. Keratinized epithelial cells transfected with sh-circ_0008450 (sh-circ_0008450) or sh-Runx3 (sh-Runx3) or both were applied for the detection of migration and EMT process. (a) Relative migratory rate was assessed by migration assay. * p < 0.05, ** p < 0.01 or *** p < 0.001. (b-c) EMT marker E-cadherin, Vimentin, Fibronectin, and ZO-1 were assessed by Western blot. ** p < 0.01 or *** p < 0.001
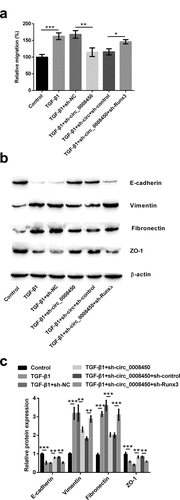
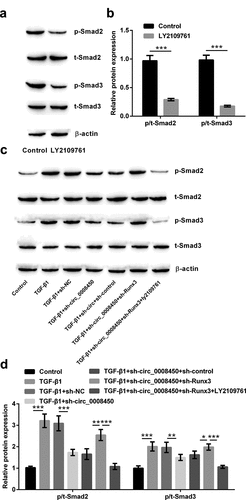
Figure 7. Circ_0008450 silence repressed the TGF-β/Smad signal pathway via the up-regulation of Runx3. (a and b) Keratinized epithelial cells treated with LY2109761 were applied for the detection of phosphorylation of Smads (Smad2 and Smad3) by Western blot. *** p < 0.001. (c and d) Keratinized epithelial cells treated with LY2109761, sh-circ_0008450 (sh-circ_0008450), sh-Runx3 (sh-Runx3) or TGF-β1 were applied for the detection of phosphorylation of Smads (Smad2 and Smad3) by western blot. * p < 0.05, ** p < 0.01 or *** p < 0.001
Data availability statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
