Figures & data
Figure 1. The expression level of HMGA2 in HUVEC under hypoxic condition
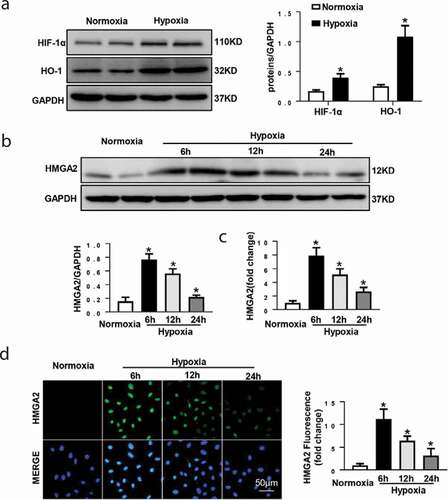
Figure 2. HMGA2 overexpression accelerated hypoxia-induced cell migration and attenuated hypoxia-induced apoptosis in HUVECs
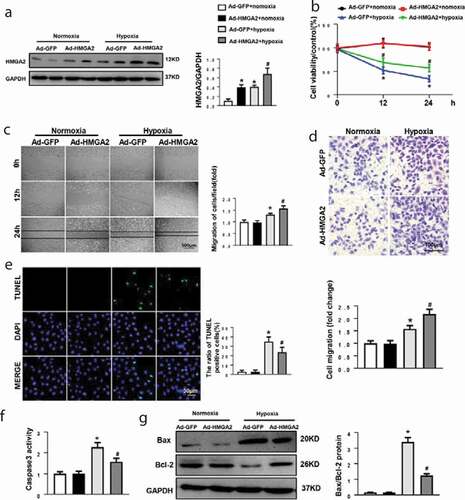
Figure 3. HMGA2 knockdown suppressed hypoxia-induced cell migration and aggravated hypoxia-induced apoptosis in HUVECs
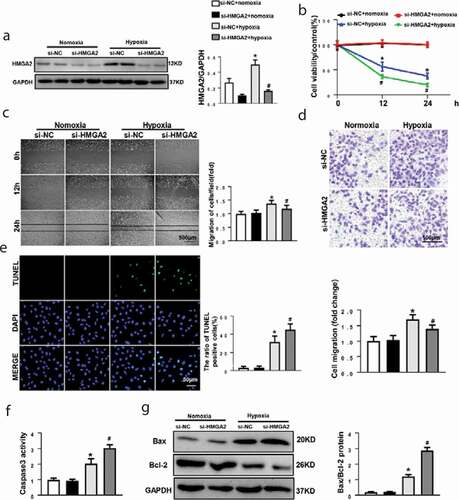
Figure 4. HMGA2 regulated HIF-1α/VEGF pathway
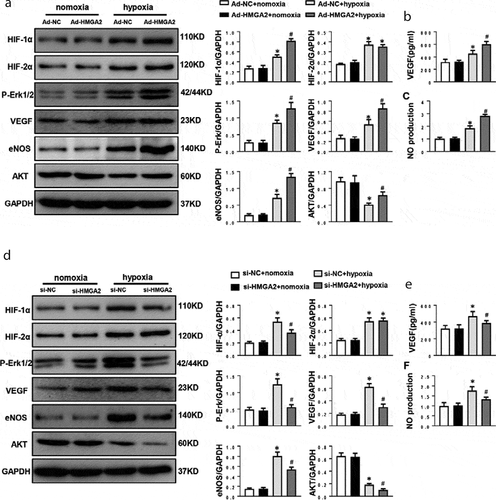
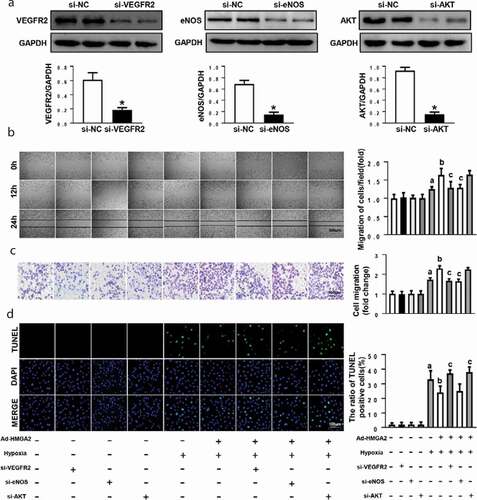
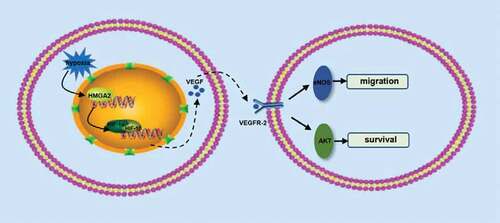
Figure 5. HMGA2 promoted migration by activating eNOS and inhibited apoptosis by regulating AKT