Figures & data
Table 1. The relationship between lncRNA GAS6-AS2 expression and the clinicopathological features in NSCLC patients
Figure 1. GAS6-AS2 was enhanced, and miR-144-3p was reduced in NSCLC tissues and cell lines. (a) qRT-PCR for GAS6-AS2 expression in NSCLC and adjacent normal tissues, n = 22. (b) The negative correlation between GAS6-AS2 and miR-144-3p in NSCLC tissues. (c and d) qRT-PCR for GAS6-AS2 and miR-144-3p expression in NSCLC cell lines (A549, H460, H1299 and H1975) compared with normal cells (BEAS-2B). *compared with adjacent normal tissues, p < 0.05; *compared with BEAS-2B, p <0.05. n = 3
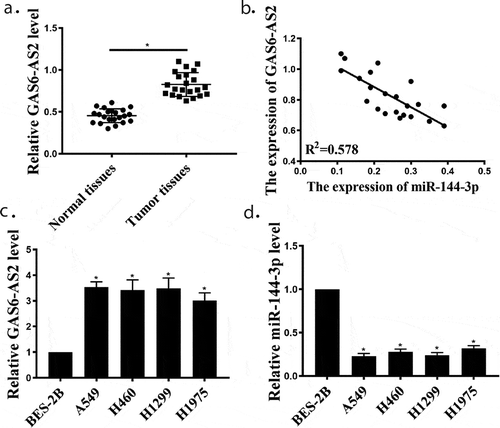
Figure 2. GAS6-AS2 promoted the growth and proliferation of A549 and H1299 cells (a) qRT-PCR for GAS6-AS2 expression in A549 and H1299 cells, transfected with GAS6-AS2 siRNA1 or GAS6-AS2 siRNA2 for 2 days. (b) Cell viabilities of A549 and H1299 cells transfected by si-GAS6-AS2-1 or si-GAS6-AS2-2. (c) Cell proliferations of A549 and H1299 cells transfected by si-GAS6-AS2-1 or si-GAS6-AS2-2. (d) Colony formation of A549 and H1299 cells transfected by si-GAS6-AS2-1 or si-GAS6-AS2-2. (e) Flow cytometry analysis of A549 and H1299 cells transfected by si-GAS6-AS2-1 or si-GAS6-AS2-2. *compared with si-NC, p <0.05
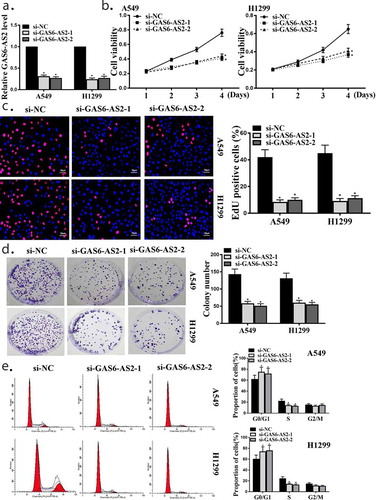
Figure 3. GAS6-AS2 functioned as a ceRNA via directly sponging of miR-144-3p. (a) Online software showed binding relationship between miR-144-3p and GAS6-AS2. (b) increased expression of miR-144-3p in miR-144-3p mimic group and reduced expression of miR-144-3p in mir-144-3p inhibitor group. (c) The co-transfection assay show the relationship of GAS6-AS2 with miR-144-3p in both A549. (d) RIP assay showed that the relationship of GAS6-AS2 with miR-144-3p in both A549. (e) RNA pull-down assay showed that the relationship of GAS6-AS2 with miR-144-3p in both A549 and H1299 cells. (f) The expression of miR-144-3p after GAS6-AS2 was silenced. *compared with NC group, p <0.05. n = 3
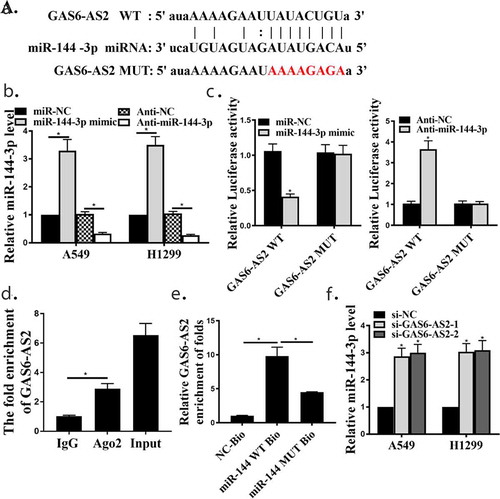
Figure 4. The relationship of miR-144-3p with cell proliferation in A549 cells. (a) CCK-8 assay showed that the relationship of miR-144-3p with cell proliferation in A549 cells. (Band C) Edu staining assay showed that the relationship of miR-144-3p with cell proliferation in A549 cells. (d and e) Colony formation assays showed that the relationship of miR-144-3p with cell proliferation in A549 cells. *compared with NC group, p <0.05. n = 3
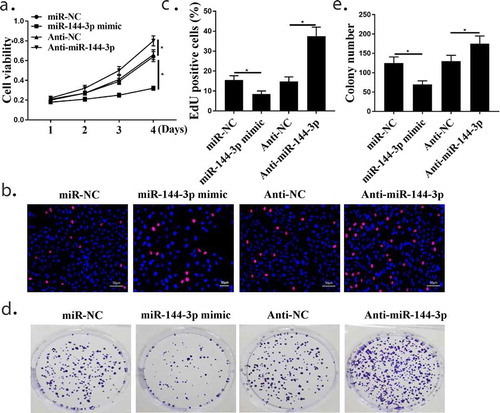
Figure 5. miR-144-3p mimic transfection weakened the influences of overexpression GAS6-AS2 in NSCLC cell proliferation. (a) qRT-PCR showed the function of pc-GAS6-AS2 in A549 and H1299 cells. (b) CCK-8 assay showed that the function GAS6-AS2 in A549 and H1299 cells. (c) Edu staining showed that the function GAS6-AS2 in A549 and H1299 cells. (d) Colony formation showed that the function GAS6-AS2 in A549 and H1299 cells. (e) (E) Flow cytometry showed that the function GAS6-AS2 in A549 and H1299 cells. *compared with NC group, p <0.05. n = 3
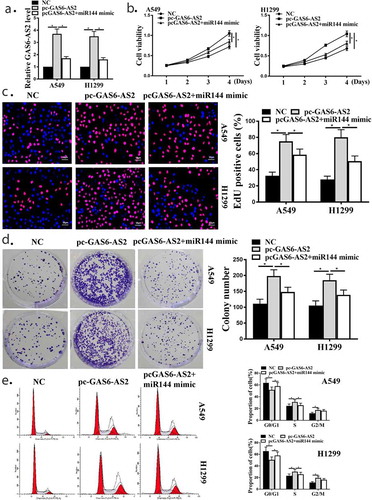
Figure 6. Relationship between MAPK6 and miR-144-3p. (a) Putative binding sites of MAPK6 and miR-144-3p are shown. (b) Luciferase activities for cells co-transfected with MAPK6-WT or MAPK6-MUT with miR-NC or miR-144-3p in A549 and H1299 cells. (c) mRNA expression of MAPK6 in A549 and H1299 cells by qRT-PCR. (d) Protein expression of MAPK6 in A549 and H1299, transfected with GAS6-AS2 or GAS6+ miR-144-3p mimic. *compared with NC group, p <0.05.n = 3
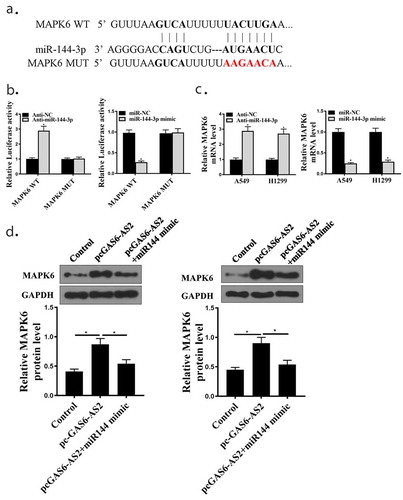
Figure 7. GAS6-AS2 promotes NSCLC cells proliferation by up-regulating MAPK6 via sponging miR-144-3p in A549 cells. (a) The transfection efficiency of MAPK6 was assessed by qRT-PCR. The cells proliferation were assessed via(b) CCK-8 assay. (c) Edu staining. (d) Colony formation. *compared with NC group, p <0.05. n = 3
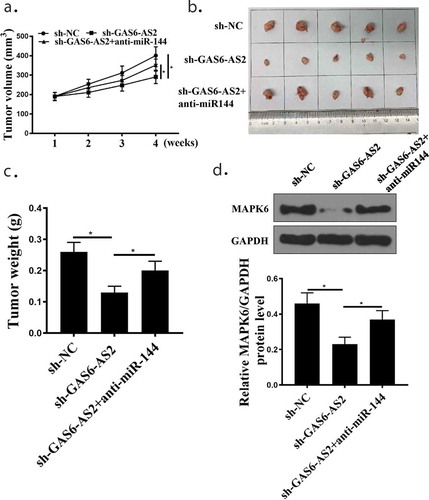
Figure 8. GAS6-AS2 silence inhibited NSCLC proliferation via miR-144-3p/MAPK6 in vivo. (a) Tumor volume was measured every week. (b) The image of tumor at the end. (c) Tumor weight was detected at the end. (d) Western blot examined the protein expression of MAPK6 in tumor tissues. *compared with NC group, p <0.05. n = 5
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
