Figures & data
Figure 1. eIF4E governs S6K1 activation state. (a and b) Expression pattern of eIF4E and S6K1 in cancer cells and tissues. Breast cancer (MCF7, MDA-MB28), Lung Cancer (A549) Colon Cancer (HCT) cells along with non-transformed HEK293, NIH3T3 (a) or representatives from Breast, Gastric, Colorectal cancer tissues (represented as T) along with their adjacent normal tissues (represented as N) (b) were harvested and analyzed by immunoblots using indicated antibodies. Quantitation represents the average of three independent experimental series normalized to actin control. Error bars denote SEM. p value denoted as * indicates P < 0.05 and ** as P < 0.01. (c and d) Phosphorylation levels of eIF4E and S6K1 in cancer tissues and Cell lines. The lysates of cell lines and tissues described in A and B, were analyzed by immunoblot as shown. To ensure that increase in the phosphorylation levels is not due to increased total protein levels in cancer samples, total protein load was optimized to bring eIF4E and S6K1 protein levels in cancer samples at par with their corresponding control samples. Unequal loading pattern is seen due to this optimization. Quantitation showing eIF4e and S6K1 phosphorylation levels represents average result of three independent experimental series normalized to their respective protein content. Error bars denote SEM. p value denoted as * indicates P < 0.05 and ** as P < 0.01. (e-g) eIF4E abundance enhances S6K1 activation. NIH3T3, HEK293 cells transfected with myc-eIF4E (e) or HEK293 cells infected with 4EBP1 shRNA (f) or eIF4E shRNA (g) were each grown in three 90 mm culture dishes. The lysate from each set was pooled such as to increase the levels of S6K1 in each lysate. The lysates were subjected to endogenous S6K1 IP to monitor S6K1 activity. S6K1 kinase activity was monitored using GST-S6 as a substrate. Immunoblots represent levels of indicated proteins
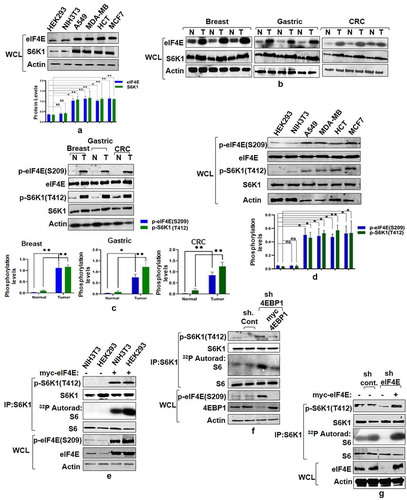
Figure 2. eIF4E phosphorylation mediates mTORC1 response onto S6K1. (a) Overexpression of eIF4E renders S6K1 rapamycin resistant. Plasmids encoding myc-eIF4E were transfected in Flag-S6K1 stable HEK293 cells in indicated manner. 60 hours post transfection, cells were either lysed directly or after treatment with 50 nM Rapamycin for 30 minutes. The lysates obtained were Flag Immunoprecipitated. S6K1 activity was monitored using GST-S6 as substrate. The immunoblots were further probed with indicated antibodies. (b) Phosphorylation state of eIF4E regulates S6K1 activity. Flag-S6K1 stable HEK293 cells lines transfected with myc-eIF4E or its Phospho mimicked (S209E) or phosphodeficient (Ser209A) were lysed directly or after 30 min incubation with 50 nM rapamycin. The lysates obtained were Flag Immunoprecipitated. S6K1 activity was monitored as described (A). The immunoblots were probed with indicated antibodies
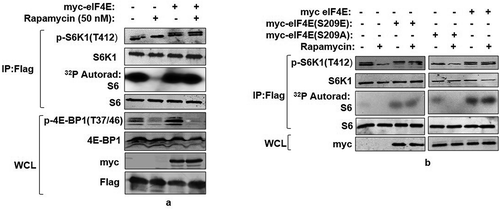
Figure 3. mTORC1 interacts with and phosphorylates eIF4E. (a and b) eIF4E phosphorylation is responsive toward mTORC1 input. Serum-stimulated Flag-S6K1 stable HEK293 cells were either lysed directly or after incubation with agents known to inhibit mTORC1 input (100 mM 2-Deoxy Glucose or 20 mM rotenone or 50 nM rapamycin for 30 minutes). Alternatively, mTORC1 inhibition was achieved by growing cells in amino acid free media (EBSS). The lysates obtained were immunoblotted and analyzed for phospho-eIF4E levels (a). HEK293 cells were infected either with mTOR or raptor shRNAs to generate respective knockdown cell lines. Scrambled shRNA was used as control. Additionally, 1 µg each of myc-mTOR and HA-raptor encoding plasmids were respectively transfected in mTOR and raptor shRNA cell lines to rescue their knock down effects. The cells were grown in serum supplemented DMEM media for 60 hours after transfection. Cells were lysed in ice-cold lysis buffer. The lysates obtained were immunoblotted and analyzed for phospho-eIF4E levels (b). (c and d) eIF4E interacts with mTORC1 adaptor protein, raptor. HA-Raptor and myc-eIF4E interact in transfected HEK293 cells. Shown are the levels of myc-eIF4E (top) and HA- Raptor and myc- tubulin (middle) in anti- HA immunoprecipitates prepared from HEK293 cells transfected with 1 µg of a plasmid encoding myc-eIF4E and 1 µg of one encoding either HA- Raptor or HA- tubulin (c). Endogenous Raptor (top) interacts with HA-eIF4E but not tubulin (bottom). Anti-HA immunoprecipitates were prepared from HEK293T cells transfected with 1 µg of either HA-eIF4E or HA-tubulin encoding plasmid (d). (e-g) eIF4E and other mTORC1 substrates interact with raptor through a novel consensus sequence motif. Representation of a novel raptor binding motif present across various mTORC1 substrates (e).Flexible protein peptide docking conducted through CABS-doc webserver represents the binding potential of the raptor with sequence motifs corresponding to ULK1, eIF4E, PRAS40 and LARP1 (f). HEK293T cells were transfected with Wild type myc-eIF4E, myc-PRAS40, HA-ULK1 and their truncation mutants, lacking potential raptor binding region. 60 hours post transfection, cells were lysed and the lysates obtained were subjected to myc and HA IP, as indicated and analyzed for endogenous raptor levels (g)
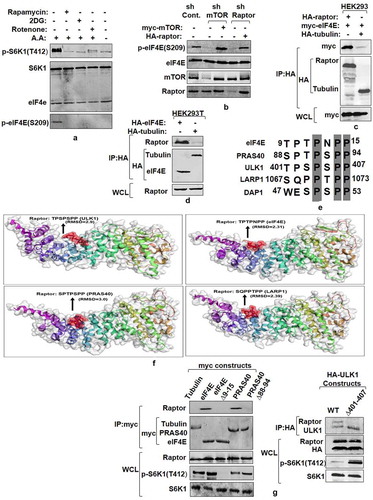
Figure 4. MNK mediated eIF4E Phosphorylation is a rapamycin-activated response (a) Assessment of MNK1 kinase activity state in cancer and non-transformed cell lines. Indicated cancer cell lines and non-transformed HEK293 and NIH3T3 cells were grown in 90 mm culture dishes, lysed and MNK1 immunoprecipitated. MNK1 kinase activity was monitored by using GST-4E as a substrate. Quantitation represents average results of three independent experimental series. Error bars denote SEM. (b) Effect of MNK1 inhibitor (CGP57380), used in indicated concentrations and for indicated time periods, was assessed on the kinase activity of MNK1, extracted from NIH3T3 and MCF7 cell lines as described in (A). Quantitation represents average results of three independent experimental series. (c, d) MNK1 is not the primary kinase for eIF4E Cell lysates obtained from indicated cell lines were treated with CGP57380 (10 µM) and rapamycin (50 nM) for 30 minutes prior to lysis. The lysates were immunoblotted and analyzed for phospho-eIF4E and phospho S6K1 levels (c). HEK293 cells were infected with MNK1 shRNAs to generate MNK1 knockdown cell line. Scrambled shRNA was used as control. Additionally, 1 µg each of HA-MNK1 encoding plasmid was transfected in MNK1shRNA cell line to rescue its knock down effect. The cells were grown in serum supplemented DMEM media for 60 hours after transfection. Cells were lysed in ice-cold lysis buffer. The lysates obtained were immunoblotted and analyzed for phospho-eIF4E levels (d). (e-f) MNK1 is a rapamycin activated eIF4E kinase. HEK293 cells were infected with control or MNK1 shRNA as described in (D). Prior to lysis, the cells were treated with 50 nM rapamycin for 12 hours. The lysates obtained were assessed for phospho-eIF4E levels (e). HEK293T cell lines were treated with 250 nM rapamycin for indicated time periods before lysis. The lysates were immunoblotted and analyzed for phospho-eIF4E and phospho S6K1 levels (f)
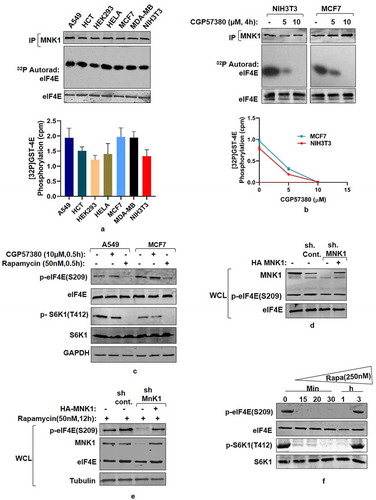
Figure 5. eIF4E interacts with S6K1-TOS motif and primes it for activation (a) eIF4E-S6K1 interaction does not depend on the activity state of S6K1. HA tagged WTS6K1, its phospho-mutant forms, S6K1T412A and S6K1T412E and Tubulin (serving as negative control) were co-transfected with myc-eIF4E in HEK293 cells. 60 hours post transfection, cells were lysed, HA immunoprecipitated and immunoblotted. The immunoblots were analyzed for eIF4E levels. (b-d) eIF4E binds TOS motif of S6K1. HA tagged WTS6K1 alongside its point mutants F28A and F16A were transfected in HEK293T cells. The cells were grown in serum supplemented DMEM for 60 hours after transfection. Cells were lysed and the lysates were subjected to HA immuno-precipitation and immunoblotting. The immunoblots were analyzed for endogenous levels of eIF4E (b).Flexible protein peptide docking conducted through CABS-doc webserver represents the binding potential of the eIF4E with TOS motifs of S6K1 and 4EBP1 (c). Flag-S6K1 stable HEK293 cells were transfected with myc-eIF4E. 60 hours post transfection, cells were lysed and lysates were subjected to myc IP. Prior to immunoblotting, increasing concentrations of intact (FDIDL) or disrupted (ADIDL) TOS motif bearing peptides were added to the immunoprecipitates in indicated manner. The immunoblots were probed for the levels of S6K1 (d). (e, f) Raptor does not mediate eIF4E-S6K1 interaction. HEK293 cells were infected with raptor shRNAs to generate raptor knockdown cell line. Scrambled shRNA was used as control. The cells were transfected with myc tagged WT-eIF4E, its phospho-mutants, and tubulin (serving as negative control). Additionally, 1 µg of HA-raptor encoding plasmid was transfected in raptor shRNA cell line to rescue its knock down effect. The cells were grown in serum supplemented DMEM for 60 hours after transfection. Cells were lysed in ice-cold lysis buffer, subjected to myc-IP and immunoblotted. The immunoblots were analyzed for S6K1 levels (e).Flag-S6K1 stable cell lines were transfected with eIF4E variant 30–217, its phosphodeficient and phosphomimicked variants. 60 hours post transfection, cells were lysed and subjected to myc-IP and immunoblotting. The immunoblots were analyzed for the levels of S6K1 (f). (g) Phosphorylation at T412 site does not compensate for TOS dysfunction. HEK293 cells were transfected in indicated manner with HA tagged WTS6K1 and ∆NHS6K1 and their phosphomimicked (T412E) variants. 60 hours post transfection, cells were lysed and subjected to HA-IP to monitor S6K1 activity. S6K1 kinase activity was monitored using GST-S6 as a substrate in an in vitro kinase assay. The immunoblot represents levels of indicated proteins. (h) eIF4E-TOS motif interaction relieves S6K1 from CTD inhibition to prime it for activation HEK293 cells transfected with HA tagged WTS6K1, TOS mutant (F28A) and carboxy terminus deleted variant of TOS mutant (F28A ∆CT), were grown in serum supplemented DMEM for 48 hours and then serum starved for 12 hours. Prior to lysis, cells were serum stimulated for 30 minutes directly or in presence of 50 nM rapamycin. The lysates were HA immunoprecipitated to monitor the S6K1 activity. The immunoblot was further analyzed for T412 phosphorylation levels of S6K1
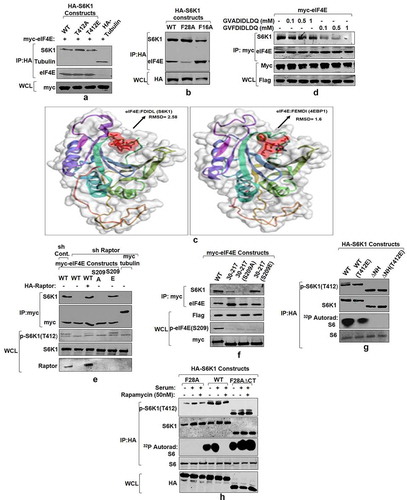
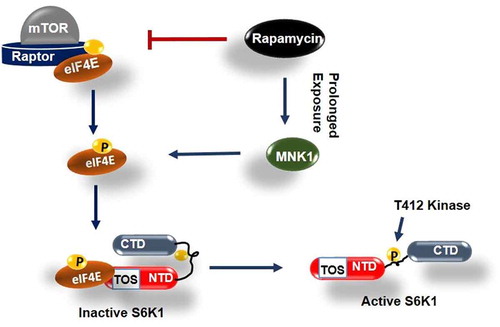
Figure 6. Model depicting role of phosphorylated eIF4E in activation of S6K1 Graphical illustration depicts that eIF4E is primarily phosphorylated by mTORC1 or alternatively by MNK1 after rapamycin activation. Phosphorylated eIF4E then interacts with TOS motif of S6K1 and relieves its inhibition due to Carboxy terminal domain (CTD). This leads to exposing of critical hydrophobic motif (HM) site of S6K1 for phosphorylation and subsequent activation by a kinase other than mTORC1