Figures & data
Table 1. Association of circ-PGC expression with clinicopathological characteristics of patients with NSCLC
Figure 1. The expression of circ-PGC and FOXR2 was elevated in NSCLC tissues and cells
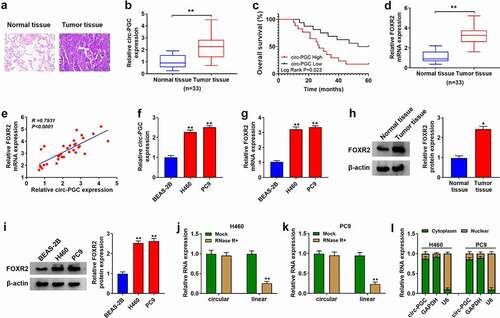
Figure 2. Circ-PGC knockdown suppressed H460 and PC9 cell malignant phenotypes
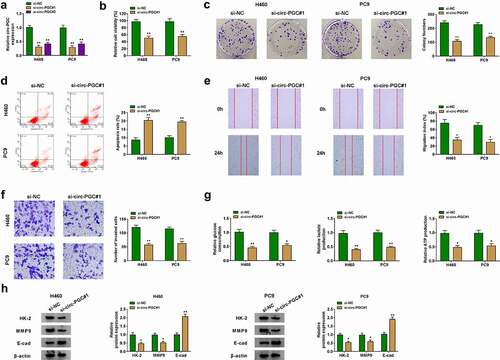
Figure 3. FOXR2 knockdown inhibited H460 and PC9 cell malignant phenotypes
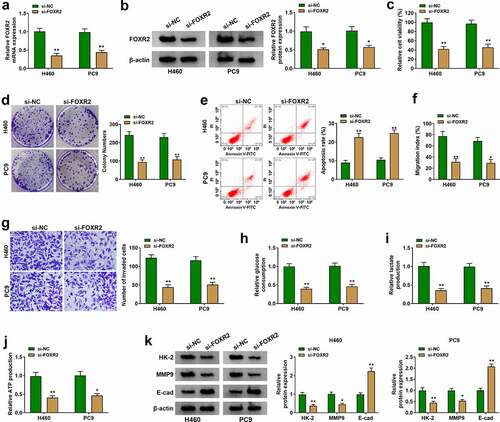
Figure 4. FOXR2 overexpression rescued the inhibitory effects on H460 and PC9 cell malignant phenotypes caused by circ-PGC knockdown
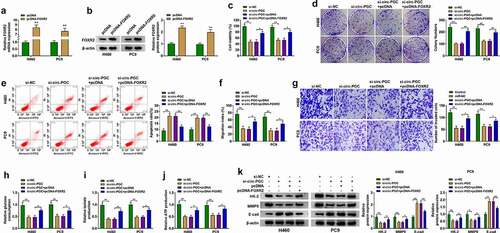
Figure 5. MiR-532-3p was a target of circ-PGC, and miR-532-3p bound to FOXR2
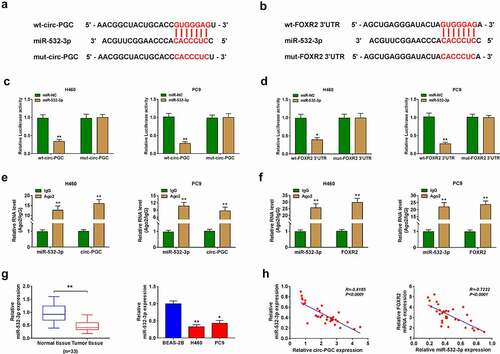
Figure 6. Circ-PGC positively regulated FOXR2 expression by targeting miR-532-3p
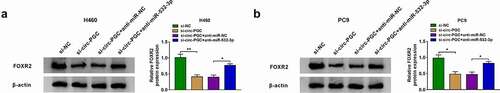
Figure 7. Wnt/β-catenin signaling pathway might be involved in the circ-PGC/miR-532-3p/FOXR2 regulatory network
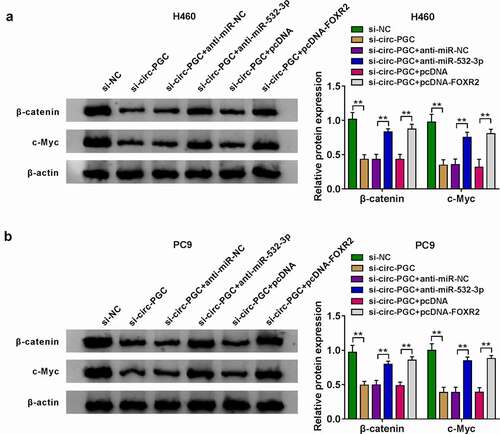
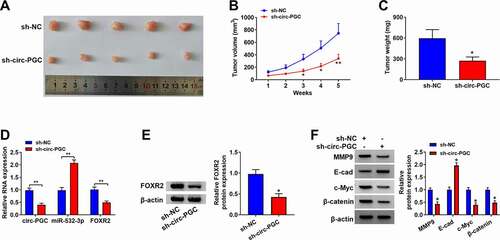
Figure 8. Circ-PGC inhibited tumor growth in vivo.
Availability of data and materials
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
