Figures & data
Table 1. List of primers used
Figure 1. miR-222-3p expression was decreased in RA-treated mouse embryos. (a) Normal and RA-treated mouse embryos at E10.5. The red arrows indicate growth retardation in the forebrain and the posterior brain. (b) qRT-PCR was used to detect the mRNA level of miR-222-3p in normal and malformed brain tissue at E8.5-E10.5. (c) miR-222-3p miRNA-Seq results
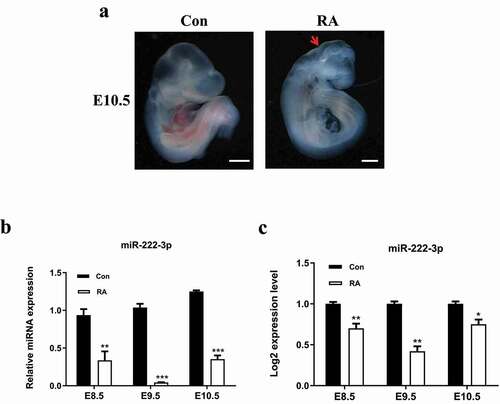
Figure 2. Inhibition of miR-222-3p affects the proliferation of HT-22 cells. (a) Transfection efficiency was detected by flow cytometry. (b) The mRNA level of miR-222-3p was analyzed by qRT-PCR in con (blank control group), anti-NC and anti-222 groups. The U6 was used as a control. (c) The proliferation of con, anti-NC and anti-222 groups cells were measured by CCK8. (d) The PCNA protein level in con, anti-NC and anti-222 groups were evaluated via western blot and the expression of β-tubulin was used for confirming equal loading
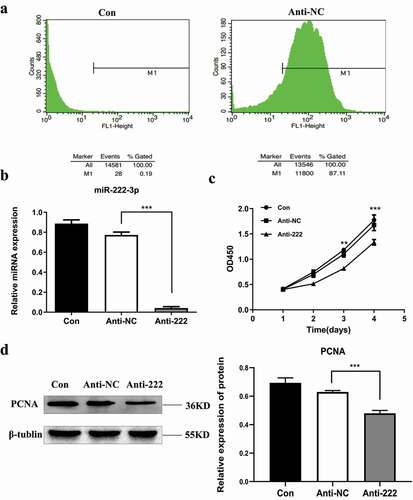
Figure 3. Inhibition of miR-222-3p affects the Cell function of HT-22 cells. (a) The cell apoptosis in con, anti-NC and anti-222 groups were analyzed by flow cytometry. (b) The Cleaved Caspase-3 protein level in con, anti-NC and anti-222 groups were evaluated via western blot and the expression of β-tubulin was used for confirming equal loading. (c) Fluorescence results of AO-EB double staining HT-22 cell nuclei. The arrows indicate apoptotic cells. (d) The cell cycle distribution in con, anti-NC and anti-222 groups was analyzed by flow cytometry. (e) The migration ability in con, anti-NC and anti-222 groups was analyzed by cell scratch experiment
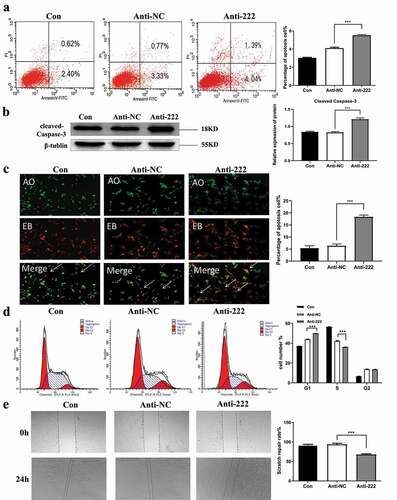
Figure 4. Ddit4 is a direct target of miR-222-3p in HT-22 cells. (a) qRT-PCR was used to detect the mRNA level of Ddit4 gene in normal and malformed brain tissue at E8.5-E10.5. (b) The mRNA level of Ddit4 gene was analyzed by qRT-PCR in con, anti-NC and anti-222 groups. The β-actin gene was used as a control. (c) The mRNA level of miR-222-3p was analyzed by qRT-PCR in con, mim-NC and mim-222 groups. The U6 was used as a control. (d) The mRNA level of Ddit4 gene was analyzed by qRT-PCR in con, mim-NC and mim-222 groups. The β-actin gene was used as a control. (e) The Ddit4 protein level in con, anti-NC and anti-222 and mim-222 groups was evaluated via western blot and the expression of β-tubulin was used for confirming equal loading. (f) Histogram analysis of graph E. (g) The fluorescence values of Ddit4 PC, Ddit4 WT and Ddit4 MUT combined with miR-222-3p mimis or NC were determined by double luciferase assay. (i) The seed sequence, mutated sequence, and complementary sequence of mir-222 and ddit4 3` UTR
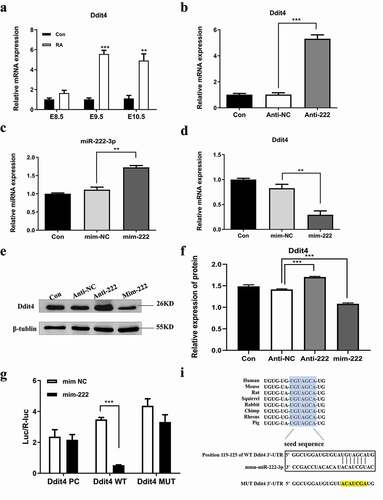
Figure 5. Ddit4 reverses the effect of miR-222-3p on HT-22 cells function. (a) The Ddit4 protein level in Con, siR-NC and siR-Ddit4 groups was evaluated via western blot and the expression of β-tubulin was used for confirming equal loading.(b)The proliferation of anti-NC, anti-222 and anti-222 + siR-Ddit4 groups cells were measured by CCK8. (c) The PCNA protein level in anti-NC, anti-222 and anti-222 + siR-Ddit4 groups was evaluated via western blot and the expression of β-tubulin was used for confirming equal loading. (d) The cell apoptosis in anti-NC, anti-222 and anti-222 + siR-Ddit4 was analyzed by flow cytometry. (e) The Cleaved Caspase-3 protein level in anti-NC, anti-222 and anti-222 + siR-Ddit4groups was evaluated via western blot and the expression of β-tubulin was used for confirming equal loading. (f)The cell cycle distribution in anti-NC, anti-222 and anti-222 + siR-Ddit4 groups was analyzed by flow cytometry. (g) The migration ability in anti-NC, anti-222 and anti-222 + siR-Ddit4 groups was analyzed by cell scratch experiment
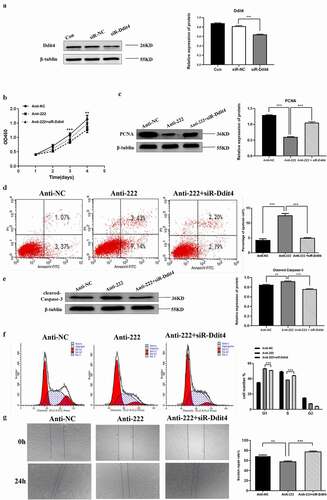
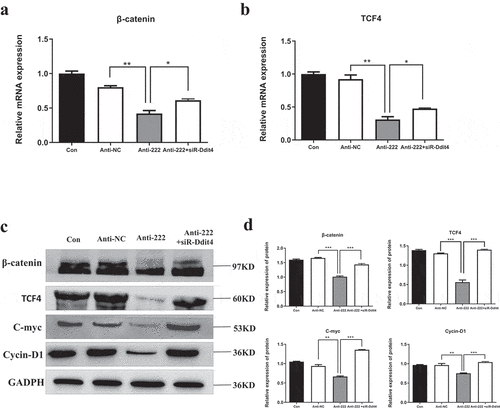
Figure 6. Inhibition of miR-222-3p regulates Wnt signaling pathway. (a) The mRNA level of β-catenin gene was analyzed by qRT-PCR in con, anti-NC, anti-222 and anti-222+ siR-Ddit4 groups. The β-actin gene was used as a control. (b) The mRNA level of TCF4 gene was analyzed by qRT-PCR in con, anti-NC, anti-222 and anti-222+ siR-Ddit4 groups. The β-actin gene was used as a control. (c) The β-catenin, TCF4, C-myc, Cycin D1 protein level in con, anti-NC, anti-222 and anti-222+ siR-Ddit4 groups was evaluated via western blot and the expression of GADPH was used for confirming equal loading