Figures & data
Figure 1. Osteogenic differentiation of bone marrow derived human MSCs. A) Alizarin Red S staining at 0, 14, 21 and 28 days of differentiation (DD) of bone marrow derived human MSCs. B) SPP1 (Osteopontin), RUNX2 and BGLAP (Osteocalcin) mRNAs relative quantification by Real Time RT-qPCR at 0, 3, 7 and 14DD. C) p53, TAp63, DNp63 and TAp73 mRNAs relative quantification by Real Time RT-qPCR at 0, 3, 7 and 14DD. Data are shown as the mean of 3 independent experiments ± SD, p-value by two-tailed unpaired Student’s t-test. D) TP63 human gene structure and description of PCR primers used in panel E) PCRs. E) Semiquantitative endpoint PCRs for p63 α, β and γ isoforms performd using 14DD MSCs cDNA
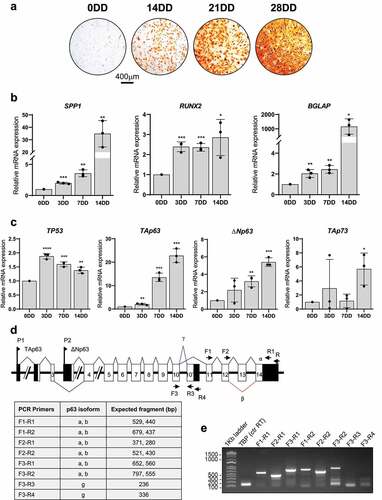
Figure 2. Osteogenic differentiation of bone marrow derived TAp63-KO (KO) e WT MSCs. A) Alizarin Red S staining at 0, 2 and 14 days of differentiation (DD) of bone marrow derived KO e WT MSC. B-C) TAp63, p53, BGLAP (osteocalcin), RUNX2 and Osterix mRNAs relative quantification by Real Time RT-qPCR at 0, 2 e 14DD in KO e WT bone marrow derived MSCs at 0, 2 e 14DD. Data are shown as the mean of 3 independent experiments ± SD, p-value by two-tailed unpaired Student’s t-test
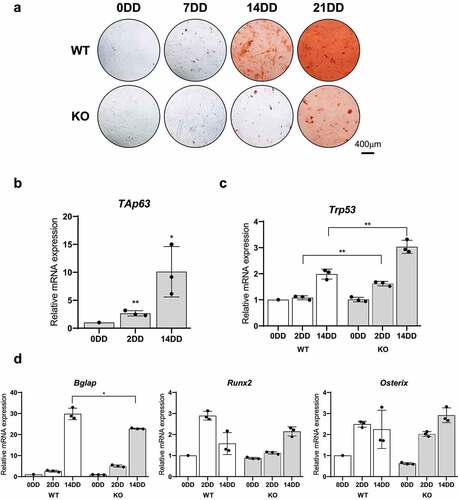
Figure 3. OPG is a TAp63 target gene during MSC osteogenic differentiation. A) OPG mRNAs relative quantification by Real Time RT-qPCR at 0, 3, 7, 14, 21 days of differentiation (DD) of bone marrow derived human MSCs. B) OPG protein levels by Western blot at 0, 3, 7, 14, 21DD of bone marrow derived human MSCs. β-actin was used as loading control. One representative experiment of three is shown. C) OPG mRNAs relative quantification by Real Time RT-qPCR at 0, 2 e 14DD in KO e WT bone marrow derived MSCs. Data are shown as the mean o 3 independent experiments ± SD, p-value by two-tailed unpaired Student’s t-test. D) TNFRSF11B fisrt intron/exon representation; the putative p63 responsive element (RE) is depicted. E) Relative luciferase activity mesured 24 h after H1299 co-transfection with pGL3-BS1 and TAp63α, TAp63β e TAp63γ expression vectors or an empty as control (Ctr). Data are shows as the mean of 4 independent experiments ± SD, p-value by two-tailed unpaired Student’s t-test. F) Western Blot on total protein extracts used in luciferase assays in E) to check HA-TAp63α, HA-TAp63β and HA-TAp63γ. *: aspecific band. β-actin was used as loading control
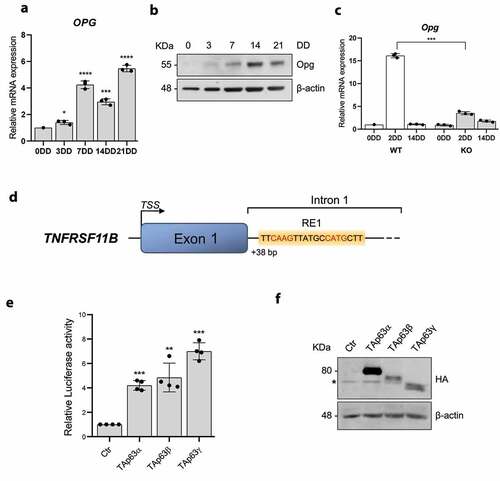
Figure 4. P63 and TNFRSF11B expression positively correlate in breast cancer patients. A) Expression of p63 and TNFRSF11B mRNA in normal, tumor and metastatic tissues. B) Bioinformatic analysis showing a positive correlation between p63 and OPG in breast cancer. C-D) High levels of both p63 and TNFRSF11B are associated with a better relapse free survival of breast cancer patients
Supplemental Material
Download Zip (619.4 KB)Data availability statement
All data generated or analysed during this study are included in this published article (and its supplementary information files). Data sharing not applicable to this article as no datasets were generated or analysed during the current study.
