Figures & data
Figure 1. RREB1 is highly expressed in gastric cancer tissue but has no significantly with the prognosis of patients
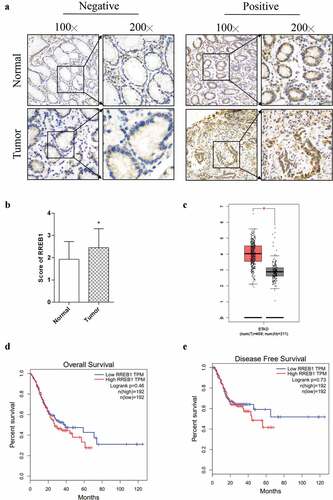
Table 1. Association between the clinicopathological features of CRC patients and RREB1 expression
Figure 2. Knockdown of RREB1 inhibit gastric cancer cell proliferation
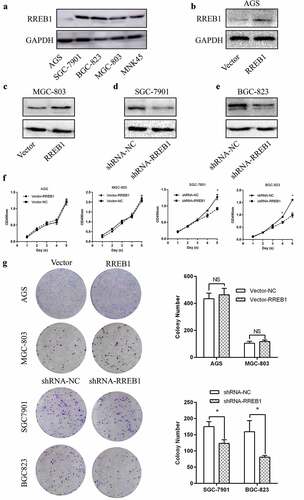
Figure 3. Cell cycle and apoptosis of RREB1 overexpression and knockdown cell lines
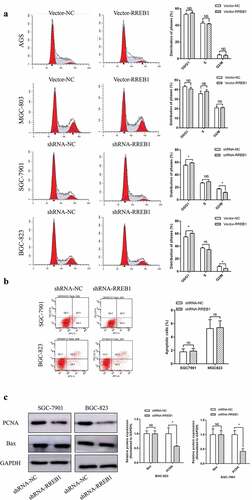
Figure 4. RREB1 promotes cell proliferation in vivo (n = 5)
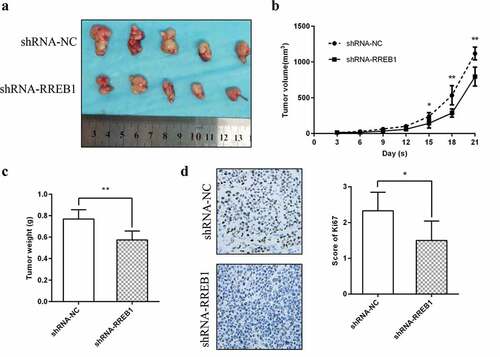
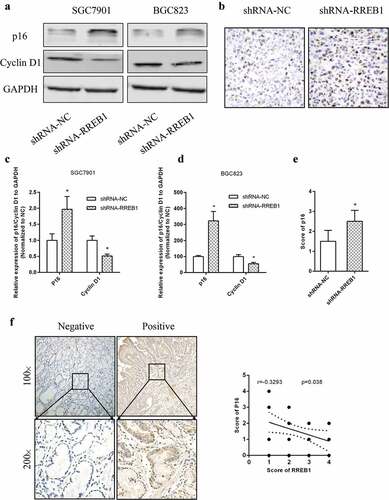
Figure 5. Knockdown of RREB1 inhibits cell proliferation via enhanced p16 expression