Figures & data
Table 1. Strains used in this work
Table 2. Primers used in this work
Figure 1. Genetic interaction between chromatin remodeler complexes and HU sensitivity. (a) Fivefold serial dilutions of the strains indicated on the left were spotted onto rich media or in media with 6 mM HU, 8 mM HU, or 10 mM HU. Plates were incubated at 30°C for 2–4 d. (b) Fivefold serial dilutions of the strains indicated on the left were spotted onto rich media or in media with 6 mM HU, 8 mM HU, or 10 mM HU. Plates were incubated at 30°C for 2–4 d
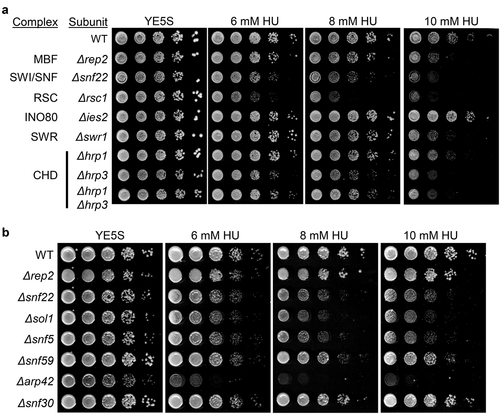
Figure 2. Snf22 binds to the promoters of MBF-regulated genes. (a) Cells expressing Snf22-Myc under the control of its own promoter were treated or not with HU for 3 h. ChIP experiments were performed using primers covering promoter (prom), coding (ORF), and termination (term) sequences of the cdc18 and cdc22 genes (as indicated). Error bars (SEM) were calculated from biological triplicates. Significant differences between treated and untreated samples were determined by the Student´s t-test (*P < 0.05). (b) Wild type (WT) or Δgcn5 cells expressing Snf22-Myc were treated or not with HU for 3 h. ChIP experiments were performed using primers covering the promoter region of the cdc18 and cdc22 genes or the SPNCRNA.650 gene as a control. Error bars (SEM) were calculated from biological triplicates. (c) Wild type (WT), Δrep2, Δyox1, or Δnrm1 cells expressing Snf22-Myc were treated or not with HU for 3 h, as indicated. Error bars (SEM) were calculated from biological triplicates. Significant differences between treated and untreated samples were determined by the Student´s t-test (*P < 0.05; ** P < 0.01)
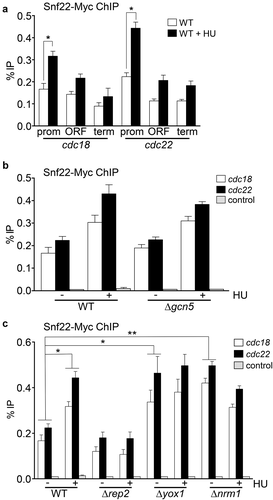
Figure 3. SWI/SNF mutant strains do not have impaired MBF-dependent transcription. (a) Expression of the MBF-dependent genes cdc18 and cdc22 was analyzed by RT-qPCR in the indicated strains, before (-) and after (+) treatment with HU for 3 h. tfb2 was used as a control for normalization. Each column represents the mean value and SEM, calculated from three biological replicates. (b) Fission yeast cdc25-22 cells or cdc25-22 Δsnf22 were synchronized at the G2/M transition. After release, samples were collected every 10 min, processed for RNA isolation, and analyzed by RT-qPCR for the expression of the genes indicated on top. Dots indicate the timepoint with the maximum value (in red, WT; in purple, Δsnf22). Error bars (SEM) were calculated from biological triplicates
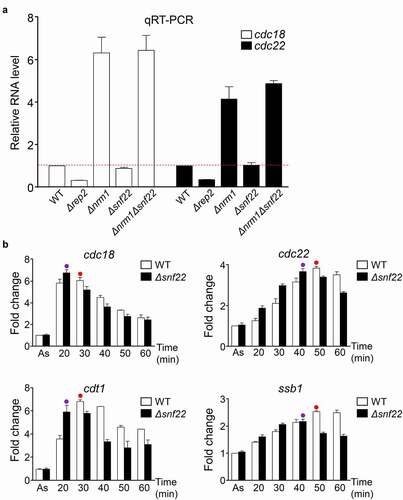
Figure 4. RSC complex mutants are sensitive to HU. (a) Fivefold serial dilutions of the strains indicated on the left were spotted onto rich media or in media with 4 mM HU, 6 mM HU, 8 mM HU, or 10 mM HU. Plates were incubated at 30°C for 2–4 d. (b) Fivefold serial dilutions of the strains indicated on the left were spotted onto rich media or in media with 4 mM HU or 6 mM HU. Plates were incubated at 25°C or 37°C, as indicated in the top
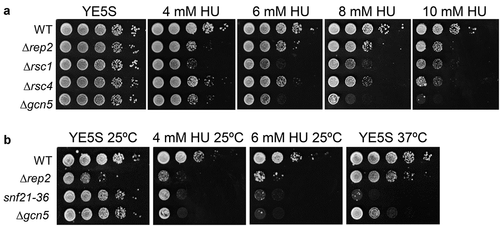
Figure 5. Snf21 binds to MBF-regulated genes. (a) Cells expressing Snf21-Myc under the control of its own promoter were treated or not with HU for 3 h. ChIP experiments were performed using primers covering promoter (prom), coding (ORF), and termination (term) sequences of the cdc18 and cdc22 genes (as indicated). Error bars (SEM) were calculated from biological triplicates. Significant differences between treated and untreated samples were determined by the Student´s t-test (*P < 0.05; **P < 0.01). (b) Wild type (WT) or Δgcn5 cells expressing Snf21-Myc were treated or not with HU for 3 h. ChIP experiments were performed using primers covering the promoter region of the cdc18 and cdc22 genes or the SPNCRNA.650 gene as a control. Error bars (SEM) were calculated from biological triplicates. Significant differences between treated and untreated samples were determined by the Student´s t-test (*P < 0.05). (c) Wild type (WT), Δrep2, Δyox1, or Δnrm1 cells expressing Snf21-Myc were treated or not with HU for 3 h, as indicated. Error bars (SEM) were calculated from biological triplicates. Significant differences between treated and untreated samples were determined by the Student´s t-test (*P < 0.05)
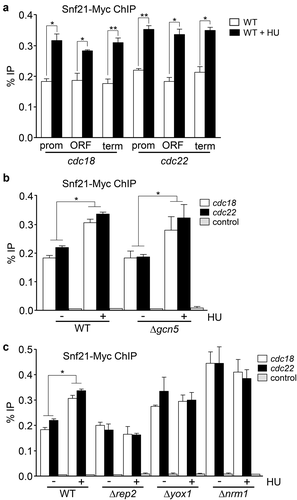
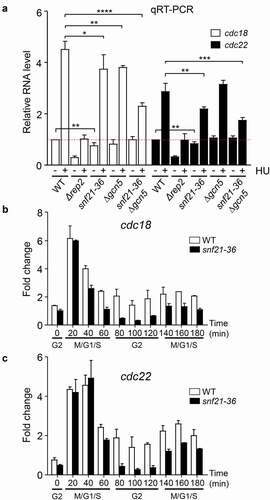
Figure 6. RSC mutant strains have impaired MBF-dependent transcription. (a) Expression of the MBF-dependent genes cdc18 and cdc22 was analyzed by RT-qPCR in the indicated strains, before (-) and after (+) treatment with HU for 3 h. tfb2 was used as a control for normalization. Each column represents the mean value and SD, calculated from three biological replicates. Significant differences between samples were determined by the Student´s t-test (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). (b) Fission yeast cdc2-asM17 cells (WT) or cdc2-asM17 snf21-36 (snf21-36) were synchronized at metaphase. After release, samples were collected every 20 min, processed for RNA isolation, and analyzed by RT-qPCR for the expression of cdc18. Error bars (SD) were calculated from biological triplicates. The different phases of the cell cycle are indicated at the bottom. (c) Same samples from (b) were analyzed for the expression of cdc22.