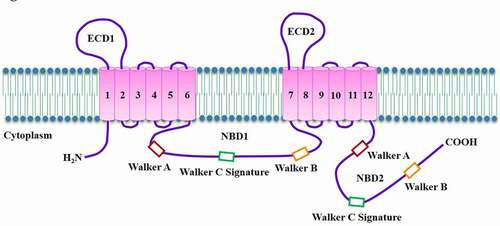
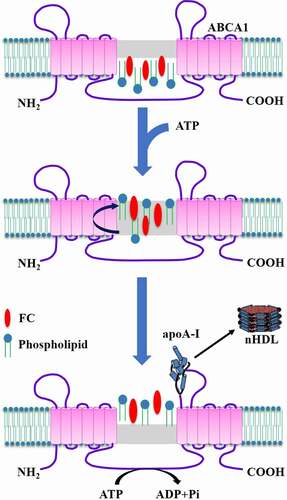
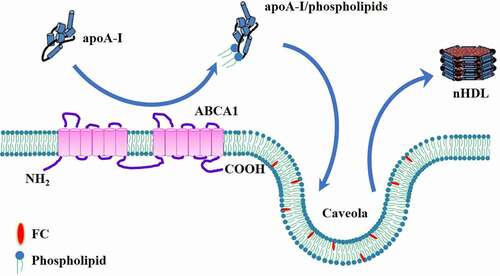
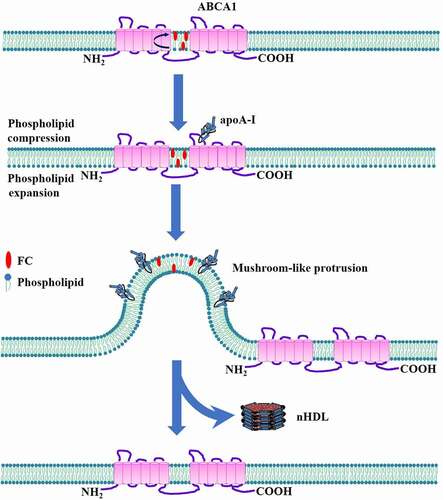
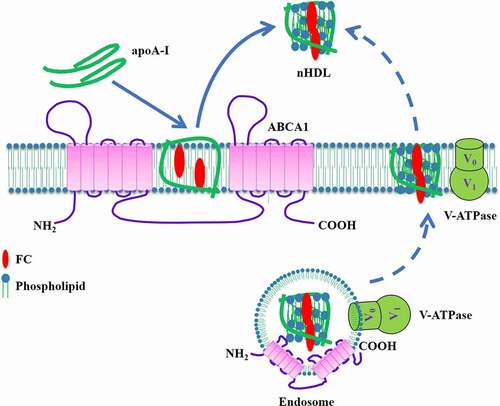
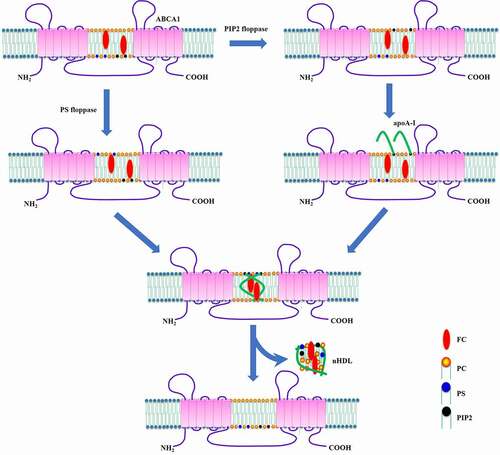
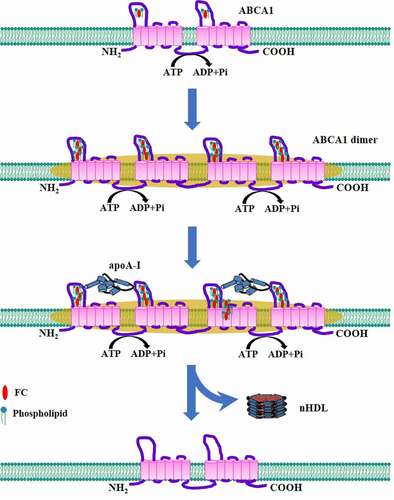
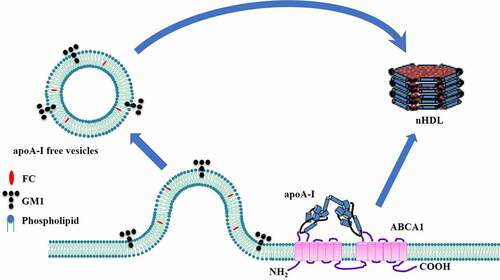
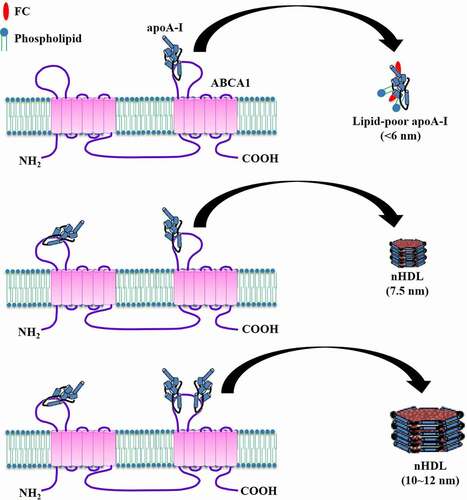
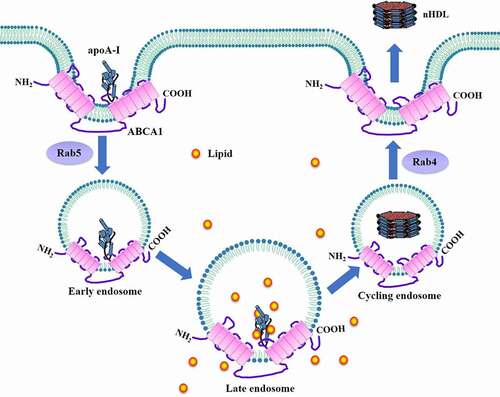
Figures & data
Table 1. The biological functions of ABCA1
Table 2. The proposed models to explain ABCA1-mediated cholesterol efflux