Figures & data
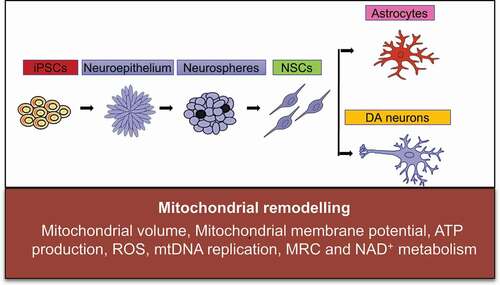
Figure 1. Mitochondrial reprogramming during the conversion of iPSCs into NSCs.
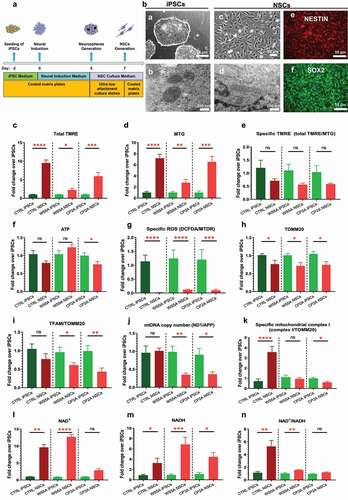
Figure 2. Mitochondrial reprogramming during the conversion of NSCs into DA neurons.
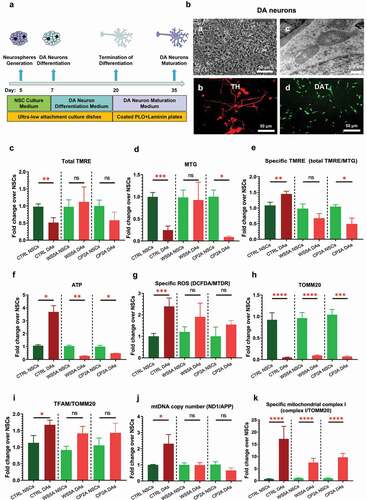
Figure 3. Mitochondrial reprogramming during the conversion of NSCs into glial astrocytes.
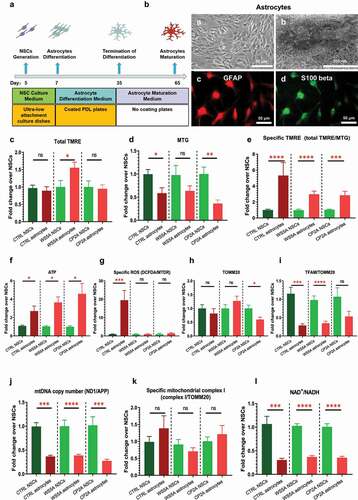
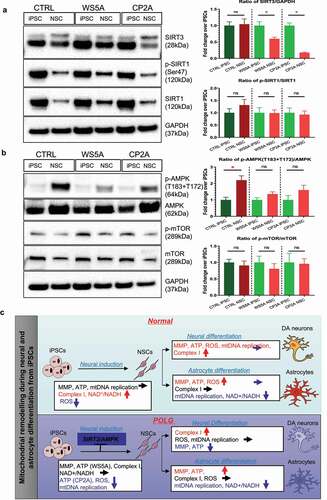
Figure 4. Abnormal mitochondrial reprogramming via downregulation of the SIRT3/AMPK signaling pathway during neural induction in POLG cells.
Supplemental Material
Download Zip (8.1 MB)Data availability statement:
We confirm that the data supporting the findings of this study are available within the supplementary materials.
