Figures & data
Table 1. Linc00883 is related to tumor typing and metastasis.
Table 2. The sequences of all primers used in qRT-PCR.
Figure 1. Expression of Linc00883 in colorectal cancer tissue samples and cells. (a) The patient cohort was obtained from the gene information of the TCGA Colon Adenocarcinoma Datasets (COAD) database that was downloaded from the cBioPortal website. The expression of Linc00883 greater than 0.87 was defined as a high expression, while the expression of Linc00883 lower than 0.87 was defined as a low expression. Kaplan-Meier analysis of the correlation between Linc00883 and survival rate of colorectal cancer (CRC) patients with low expression of Linc00883 (n = 189) and with high expression of Linc00883 (n = 187). (b) Quantitative Real-Time PCR (qRT-PCR) was applied to detect the expression of Linc00883 in patients with distant metastasis of CRC (M1: n = 29) or non-metastatic CRC (M0: n = 199). The expression of Linc00883 in CRC tissues (n = 64) (c) and CRC cell lines (d) was assessed using qRT-PCR. ***P < 0.001 vs. Peri-tumor or NCM460 (colon epithelial cell line). All experiments were performed at least in triplicate.
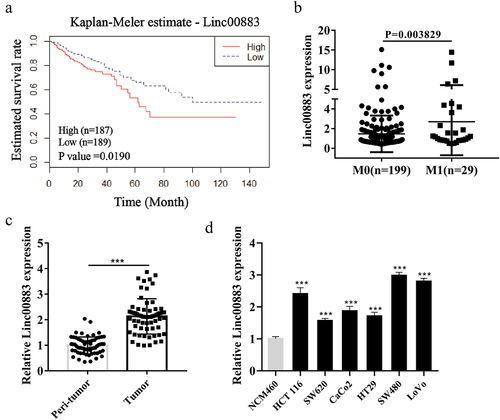
Figure 2. Influence of Linc00883 on CRC cell proliferation, invasion, and migration. Si-Linc00883 and its corresponding control were transfected into CRC cell lines SW480 and LoVo. (a) Analysis of the Linc00883 expression in CRC cells by qRT-PCR. (b) Cell Counting Kit-8 (CCK-8) was applied to analyze CRC cell proliferation. (c-d) The CRC cell invasion was assessed using a transwell assay (Scale bar: 100 μm). (e-f) A wound-healing experiment was carried out to detect CRC cell migration (Scale bar: 200 μm). **P < 0.01, ***P < 0.001 vs. si-NC. NC: Negative control. All experiments were performed at least in triplicate.
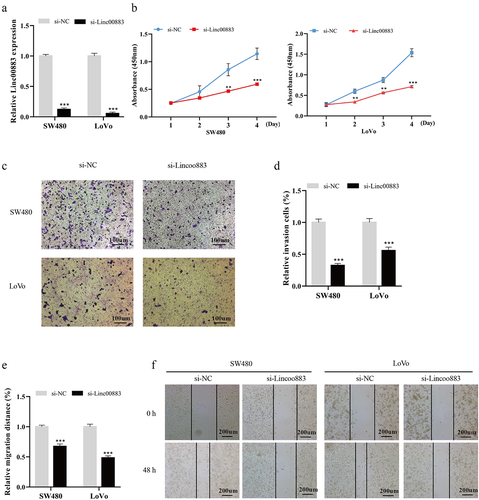
Figure 3. Effect of Linc00883 on miR-577 expression. (a) The Linc00883 expression in the nucleus or cytoplasm of cells was measured using qRT-PCR. (b) qRT-PCR was applied to detect miR-577 expression in CRC tissue samples (n = 64) and cell lines. (c) The correlation analysis between Linc00883 and miR-577. (d) The online bioinformatics (microRNA.org) was performed to predict the binding sites between Linc00883 and miR-577. The si-Linc00883, pcDNA-Linc00883, and their corresponding controls were transfected into SW480 and LoVo cells. (e) Analysis of the miR-577 expression by qRT-PCR. (f) A dual-luciferase reporter gene assay was conducted to assess the influence of miR-577 on Linc00883 luciferase activity. (g-h) RIP and RNA pull-down assays were performed. **P < 0.01 vs. lgG or Bio-NC; ***P < 0.001 vs. nucleus, Peri-tumor, NCM460, si-NC, pcDNA-NC or mimics-NC. All experiments were performed at least in triplicate.
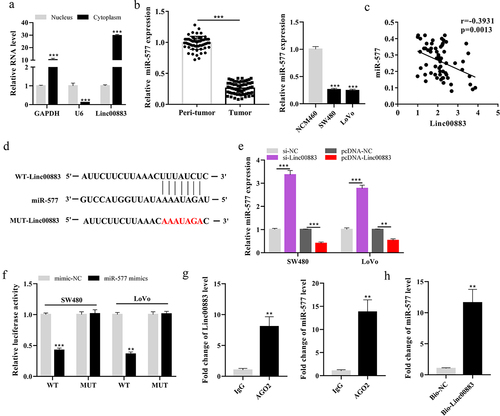
Figure 4. Linc00883 influences CRC cell proliferation, invasion, and migration via miR-577. The pcDNA-Linc00883 or/and miR-577 mimics and their corresponding controls were transfected into SW480 and LoVo cells. (a-b) The expressions of Linc00883 and miR-577 were measured using qRT-PCR. (c-d) CCK-8 was applied to assess CRC cell proliferation. (e-f) The CRC cell invasion was measured using transwell assay (Scale bar: 100 μm). (g-i) A wound-healing experiment was performed to assess CRC cell migration (Scale bar: 200 μm). *P < 0.05, **P < 0.01, ***P < 0.001 vs. pcDNA-NC + mimics-NC. #P < 0.05, ##P < 0.01 vs. pcDNA-NC + miR-577 mimics. NC: Negative control. All experiments were performed at least in triplicate.
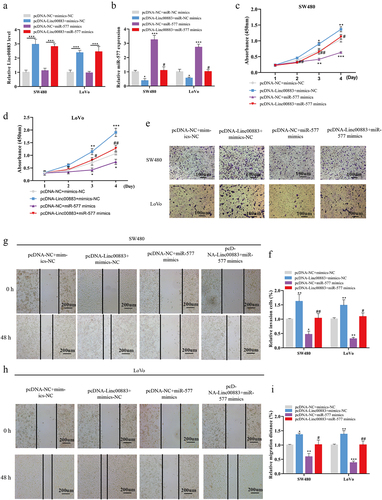
Figure 5. Linc00883 regulates CRC cell proliferation, invasion, and migration via the miR-577/FKBP14 axis. (a) Bioinformatics online software (StarBase) was applied to forecast the potential binding sites between miR-577 and FKBP14. TCGA database was applied to analyze the expression in colon cancer (B, Normal: n = 41 and Primary tumor: n = 286) or the effect of FKBP14 on the prognosis of colon cancer (C, high expression FKBP14: n = 70; low/medium expression FKBP14: n = 209). (d) The protein level of FKBP14 in CRC tissue samples was detected by western blot. (e) The mRNA level of FKBP14 in CRC tissue samples was detected by qRT-PCR. (f) The correlation analysis between FKBP14 and miR-577. The si-Linc00883 or/and miR-577 inhibitor were transfected into SW480 cells. (g-h) The expressions of Linc00883 and miR-577 were detected by qRT-PCR. (i) Detection of FKBP14 mRNA and protein levels using qRT-PCR and western blot. The pcDNA-Linc00883 or/and miR-577 mimics was transfected into SW480 cells. (j) Detection of FKBP14 mRNA and protein levels by qRT-PCR and western blot. (k) Detection of the luciferase activity of FKBP14 using dual-luciferase reporter gene assay. The si-FKBP14 was transfected into SW480 and LoVo cells. (l) The CRC cell proliferation was assessed by CCK-8. (m) Transwell assay was applied to analyze CRC cell invasion (Scale bar: 100 μm). (n) The CRC cell migration was assessed using a wound-healing experiment (Scale bar: 200 μm). *P < 0.05, **P < 0.01, ***P < 0.001 . All experiments were performed at least in triplicate.
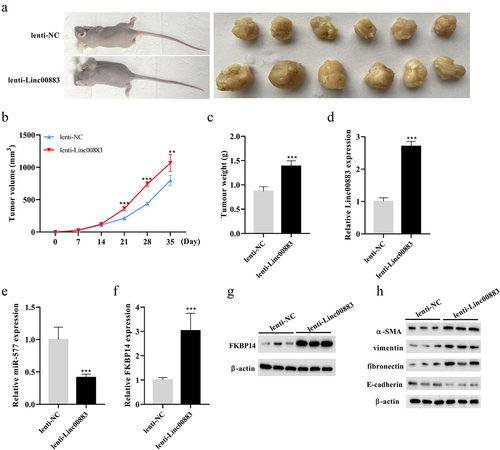
Figure 6. Effect of Linc00883 on CRC tumor growth in vivo. A xenograft nude mouse model of CRC by subcutaneously injecting CRC cells SW480 transfected with lenti-Linc00883 was established. n = 6. (a-b) Detection of the volume of CRC tumors. (c) Detection of the weight of CRC tumors. (d-f) Detection of Linc00883, miR-577, and FKBP14 expressions by qRT-PCR. (g) Detection of the FKBP14 protein level using western blot. (h) Detection of the protein levels of epithelial-mesenchymal transition (EMT)-related proteins E-cadherin, vimentin, fibronectin, and α-SMA by western blot. **P < 0.01, ***P < 0.001 vs. lenti-NC.