Figures & data
Figure 1. The miR-206/CREB5/PI3K/AKT axis might be involved in HCC growth and metastasis. A, GO analysis of DEGs in GSE49515 dataset. The three histograms in the figure indicate the enrichment of BP, CC, and MF in the GO analysis. The abscissa indicates the enrichment items, and the ordinate indicates the number of genes. B, Heatmaps of the top 30 DEGs in HCC of the GSE49515 dataset (n = 10 for normal control sample; n = 10 for HCC sample). C, Network diagram of the interaction of the top 30 DEGs. D, Prediction of the interaction between mmu-miR-206 and the CREB5 gene through TargetScan. E, The expression of 10 miRNAs regulating CREB5 in HCC was screened out by RT-qPCR, n = 10. * p < 0.05 vs. the control.
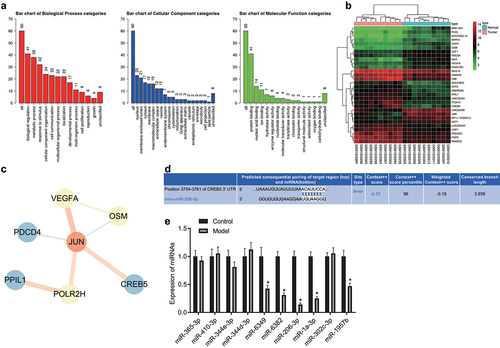
Figure 2. CREB5 overexpressed in tissues from HCC mice. A, Livers of HCC mice obtained by dissection. B, HE staining results of liver tissues from control and HCC mice (× 400). C, Positive expression of CREB5 protein in liver tissues from control and HCC mice, as determined by immunohistochemical staining (× 400). * p < 0.05 vs. the control mice. n = 10 mice in each group.
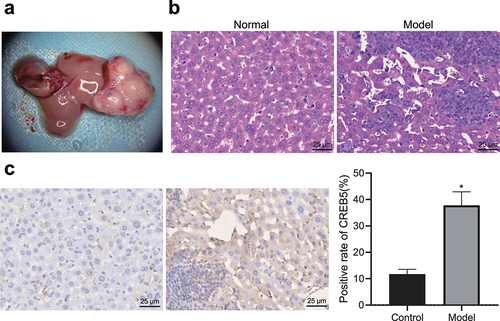
Figure 3. miR-206 targets CREB5 to suppress PI3K/AKT signaling pathway. A, Targeting relationship between miR-206 and CREB5, as assessed by dual-luciferase reporter gene assay. B, RT-qPCR detection of the expression of miR-206, CREB5, and PI3K/AKT signaling pathway-related factors in MHCC97-H cells transfected with miR-206 mimic or oe-CREB5. C, Western blot analysis of protein expression of CREB5 and PI3K/AKT signaling pathway-related factors in MHCC97-H cells transfected with miR-206 mimic or oe-CREB5. * p < 0.05 vs. the NC treatment. # p < 0.05 vs. MHCC97-H cells transfected with miR-206 mimic. Cell experiments were repeated for three times.
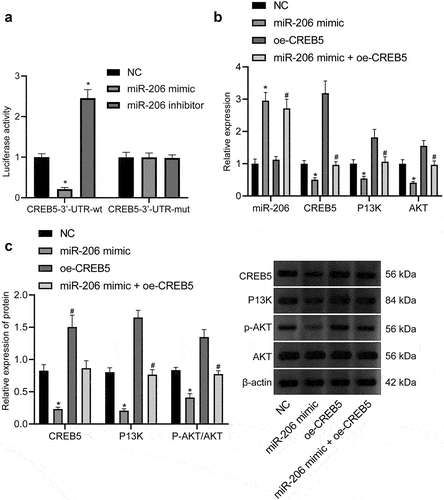
Figure 4. miR-206 overexpression blocks the PI3K/AKT signaling pathway and depresses proliferation, migration, and invasion but accelerates apoptosis in HCC cells. MHCC97-H cells were treated with miR-206 mimic, miR-206 inhibitor, NC, Wortmannin, miR-206 inhibitor + sh-CREB5, or miR-206 inhibitor + Wortmannin. A, RT-qPCR detection of the expression of miR-206, CREB5, and PI3K/AKT signaling pathway-related factors in MHCC97-H cells. B, Western blot analysis of protein expression of CREB5 and PI3K/AKT signaling pathway-related factors in MHCC97-H cells. C, MHCC97-H cell viability was assessed by MTT assay. D, Colony formation of MHCC97-H cells was assessed by plate colony formation assay. E, MHCC97-H cell migration was detected by scratch test. F, MHCC97-H cell invasion was detected by Transwell assay. G, MHCC97-H cell apoptosis was evaluated by flow cytometry. * p < 0.05 vs. MHCC97-H cells treated with NC. # p < 0.05 vs. MHCC97-H cells treated with miR-206 inhibitor. The cell experiments were repeated 3 times.
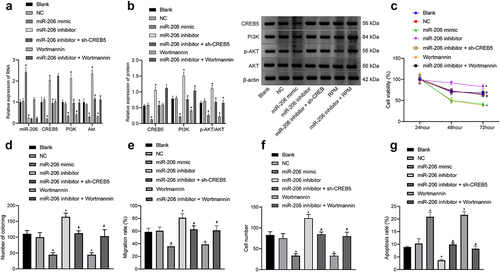
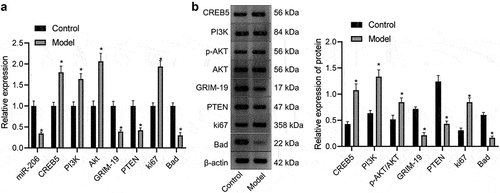
Figure 5. Expression of miR-206, Bad, GRIM-19 and PTEN is reduced, but expression of PI3K, AKT, Ki67, and CREB5 is enhanced in HCC mice. A, miR-206 expression and Bad, GRIM-19, PTEN, PI3K, AKT, Ki67, and CREB5 mRNA expression in hepatocytes of control and HCC mice, as evaluated by RT-qPCR. B, western blot analysis of Bad, GRIM-19, PTEN, PI3K, AKT, Ki67, and CREB5 protein expression in the liver tissues of control and HCC mice. * p < 0.05 vs. the control mice. n = 20 mice in each group. For this measurement data, data are shown as the mean ± standard deviation, and an independent sample t test was used for comparisons between two groups.