Figures & data
Figure 1. DEX inhibits VSMC phenotype switch. (a-b) BrdU incorporation assay for assessing VSMC proliferation in each group. (c-f) Wound healing and Transwell assays for evaluating VSMC migratory ability in each group. (g-h) Western blotting for examining protein levels of contractile markers (α-SMA and SM-22α) and synthetic markers (vimentin, osteopontin). (i-j) ELISA for measuring production of cytokines (TNF-α, IL-6) in VSMCs. Each experiment was performed in triplicate.Each experiment was performed in triplicate. **p˂0.01 vs. control+ DMSO group; #p˂0.05 vs. Ang II+ DMSO group; &p˂0.05 vs. control+ DEX group.
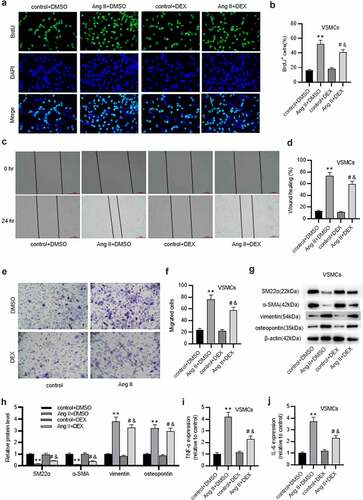
Figure 2. DEX protects against ECM degradation in VSMCs. A-C. Western blotting of collagen protein levels in VSMCs of each group. D-F. Western blotting of MMP-2 and MMP-9 protein levels in VSMCs of each group. G-I. Gelatin zymography assay for detecting MMP-2 and MMP-9 activities in VSMCs. Each experiment was performed in triplicate. **p˂0.01 vs. control+ DMSO group; #p˂0.05 vs. Ang II+ DMSO group; &p˂0.05 vs. control+ DEX group.
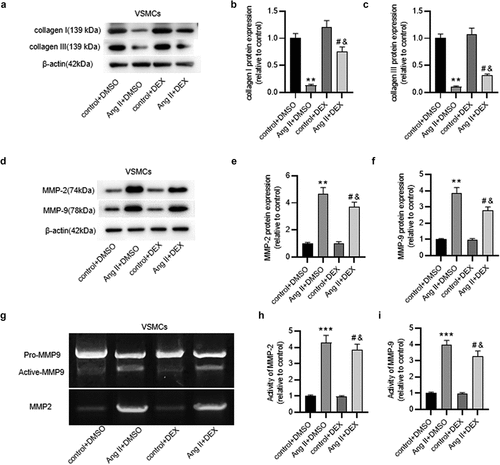
Figure 3. DEX attenuates inflammation in Ang II-treated ECs. (a) if staining for detecting ICAM-1 expression in ECs. (b) Quantification of ICAM-1 expression in each group. (c) Adhesion of THP-1 monocytes to ECs observed under a light microscope. (d) Quantification of THP-1 cell adhesion in each group. Each experiment was performed in triplicate. **p˂0.01 vs. control+ DMSO group; #p˂0.05 vs. Ang II+ DMSO group; &p˂0.05 vs. control+ DEX group.
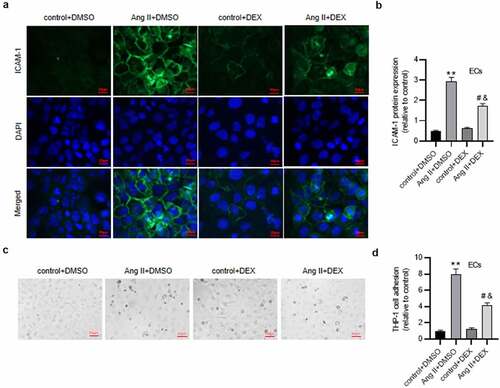
Figure 4. DEX alleviates the apoptosis of ECs. (a-b) Flow cytometry analysis for detecting EC apoptosis in each group. Quadrants 2 and 3 (Q2, Q3) were considered as apoptotic cells. (c) Western blotting for measuring levels of apoptosis-associated proteins in ECs. Each experiment was performed in triplicate. *p˂0.05, **p˂0.01.
Figure 5. DEX suppresses the expression of HMGB1, TLR4 and p-p65 in ECs. (a-b) Western blotting for analyzing protein levels of HMGB1, TLR4, p-p65 and NF-κB p65 in ECs of each group. Each experiment was performed in triplicate. *p˂0.05, **p˂0.01.
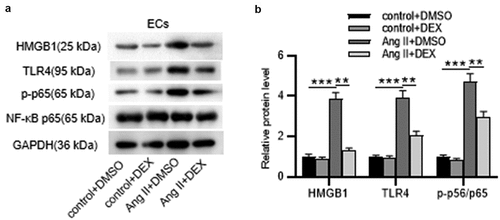
Figure 6. DEX affects the inflammation and apoptosis of ECs via HMGB1/TLR4/NF-κB signaling pathway. (a-b) Western blotting for assessing HMGB1/TLR4/NF-κB signaling pathway-related proteins in ECs. (c) Adhesion of THP-1 monocytes to ECs observed under a light microscope. (d) Quantification of THP-1 cell adhesion in each group. (e) Western blotting measuring levels of apoptosis-associated proteins in ECs. Each experiment was performed in triplicate. ***p˂0.001 vs. control+ DMSO group; #p˂0.05 vs. Ang II+ DMSO group.
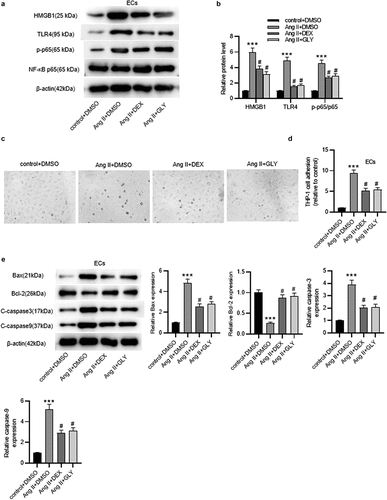
Figure 7. DEX attenuates Ang II-induced AAA and inflammation in mice. (a) Representative images of HE staining for histopathological observation of murine abdominal aortic tissue. (b-c) Abdominal aortic diameter and media/lumen ratio of each group. (d-f) ELISA for determining the concentrations of proinflammatory cytokines in murine abdominal aortic samples. n = 10 mice/group. Each experiment was performed in triplicate. **p˂0.01 vs. control+ DMSO group; #p˂0.05, &p˂0.05vs. Ang II+ DMSO group.
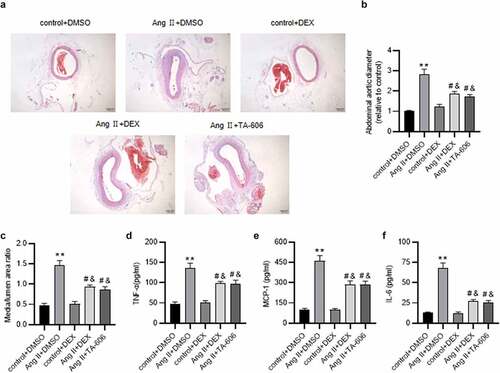
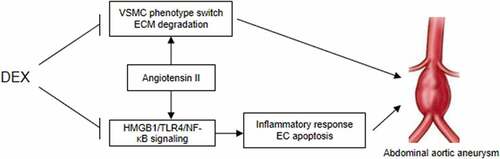
Figure 8. A schematic view of the in vitro model investigated in this study. VSMCs and ECs were treated with Ang II to mimic the pathological condition of AAA in vitro. DEX can suppress Ang II-evoked VSMC phenotype switch and ECM degradation, and it inactivates the HMGB1/TLR4/NF-κB signaling pathway to attenuate inflammatory response and the apoptosis of ECs, thereby ameliorating the condition of AAA.
Supplemental Material
Download MS Word (31.5 KB)Supplemental Material
Download MS Word (31.3 KB)Data availability statement
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.
