Figures & data
Figure 1. Schematic representation of the most common and best studied methyl-based modifications of the various classes of RNA, along with their main human methylases (“writers”), demethylases (“erasers”) and “readers”, which drive cellular responses when methylated ribonucleotides are detected. ALKBHs, AlkB Homolog 1, histone H2A dioxygenase; ALYREF, Aly/REF export factor; CMTR1–2, Cap methyltransferases 1 and 2; DNMT, DNA methyltransferases; FBL, fibrillarin; FTO, Fat mass and obesity-associated protein; FTSJ1–2, FtsJ RNA 2’-O-methyltransferases 1 and 2; HENMT1, HEN methyltransferase 1; IGF2BPs, insulin-like growth factor 2 mRNA binding proteins; METTLs, methyltransferase complex subunits; MRM1–3, mitochondrial rRNA methyltransferases 1–3; NML, nucleomethylin; NSUN, NOP2/Sun RNA methyltransferases; PCIF1, phosphorylated CTD interacting factor 1; RNMT, RNA guanine-7 methyltransferase; TRMT, tRNA methyltransferase; WTAP, Wilms tumor 1-associating protein; YBX1, Y-Box Binding Protein 1; YTHDCs, YTH Domain Containing proteins; YTHDFs, YTH N6-methyladenosine RNA binding proteins; ZCCHC4, zinc finger CCHC-type containing 4.
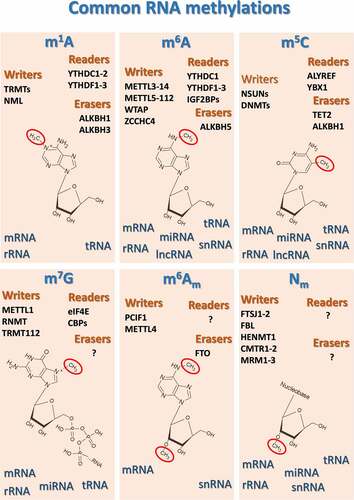
Table 1. RNA methylation and cancer resistance traits potentially related to redox rewiring.
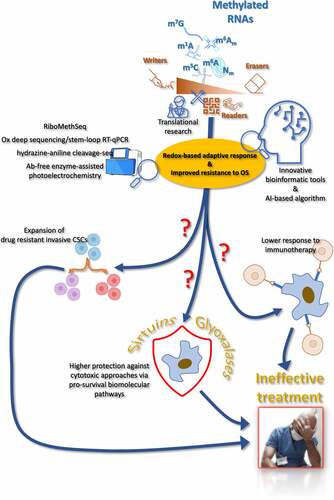
Figure 2. Schematic representation of the possible directions that future research activities and efforts may take to reveal in detail the role of the methylepitranscriptome in affecting or determining cancer cell response to treatments, also in terms of potential participation of redox-sensitive and redox-dependent biomolecular mechanisms.