Figures & data
Figure 1. Phosphorylation of CypA at Ser77 regulates subcellular localization.
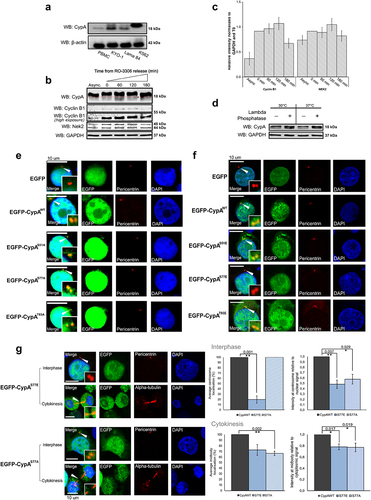
Figure 2. CypA interacts with Nek2 and PP1 during the cell cycle.
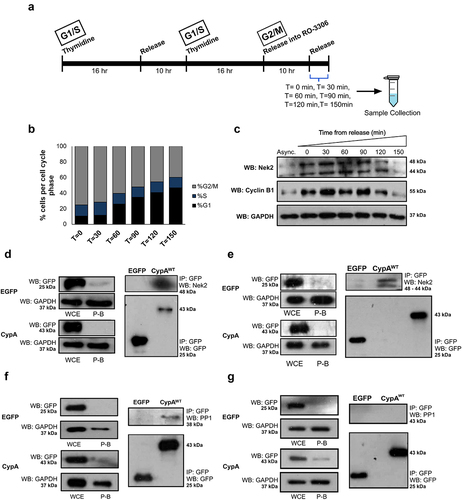
Figure 3. CypA is phosphorylated by Nek2 in vitro.
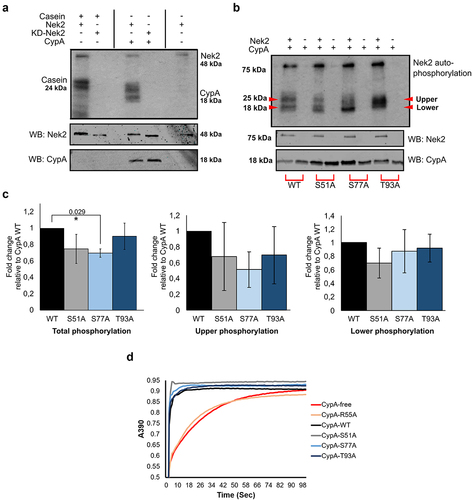
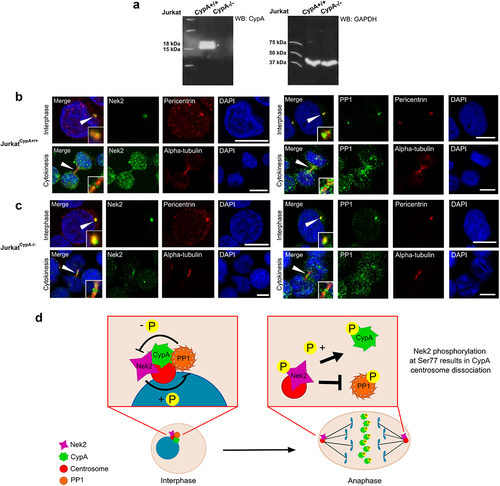
Figure 4. CypA does not affect Nek2 or PP1 localization to the midbody.
Supplemental Material
Download Zip (22.6 MB)Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article and/or its supplementary materials.
