Figures & data
Figure 1. Metformin impedes MM cell proliferation and promotes apoptosis.
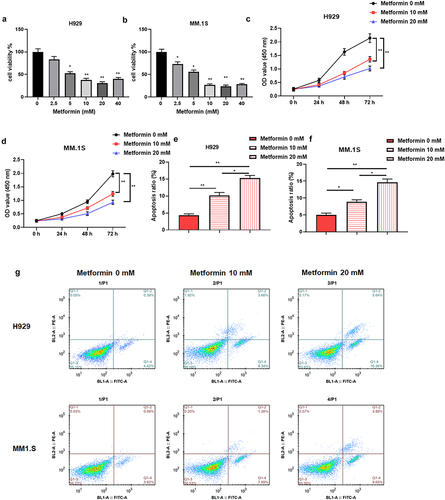
Figure 2. Metformin was able to reduce m6A abundance in MM.
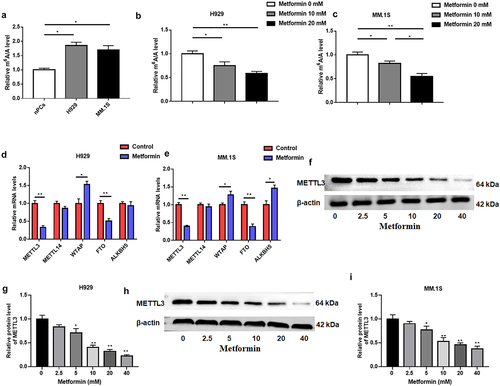
Figure 3. METTL3 overexpression counteracts metformin-mediated suppression of MM cell proliferation and facilitative effect of cell apoptosis.
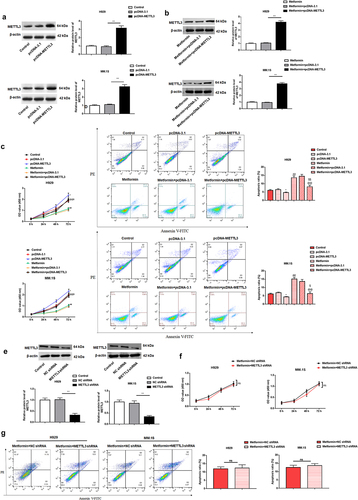
Figure 4. Mechanistic excavation into the involvement of metformin in MM progression through regulation of METTL3.
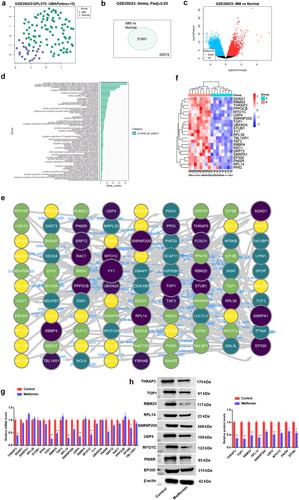
Figure 5. Metformin weakened METTL3-mediated m6A methylation of THRAP3, RBM25 and USP4.
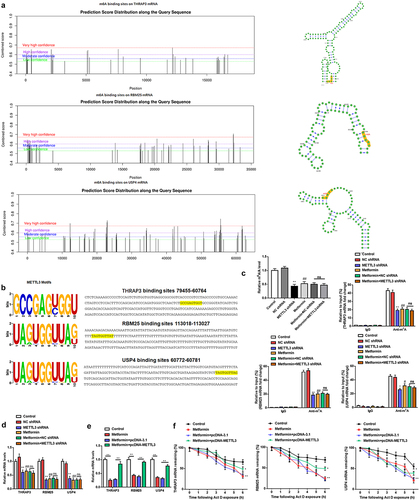
Figure 6. THRAP3, RBM25 or USP4 reversed the assistance of METTL3 on the malignant behavior of MM cells.
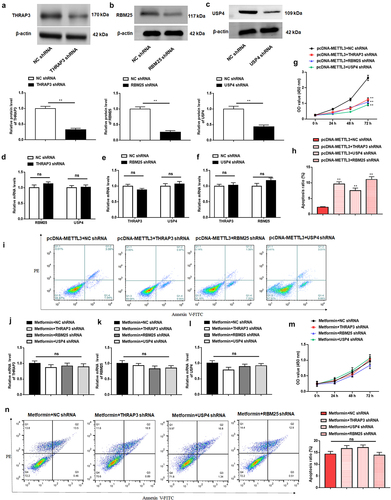
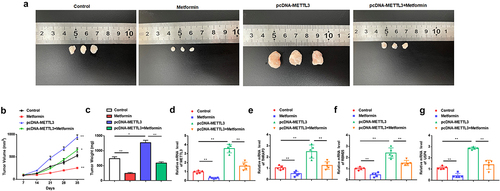
Figure 7. Metformin inhibits MM xenograft tumor growth by repressing METTL3 expression in nude mice.