Figures & data
Table 1 Demographics and lung function
Table 2 Fractionation conditions
Figure 1 Differences in IMAC protein chip profile between a COPD patient and a well matched control. (a) A 52-year-old, 108 pack-year man with COPD (FEV1 = 53% predicted) and (b) a 52-year-old, 105 pack-year man with normal airflow. The arrows demonstrate that the intensity of the protein with m/z peak at 7961.19 Da is noticeably lower in the patient with COPD. There were no visible differences in other peaks such as that of 7794.57 Da. The x-axis represents the m/z ratio from 7750–8100 Da. The y-axis shows peak intensities normalized for total ion current and corrected with baseline subtraction.
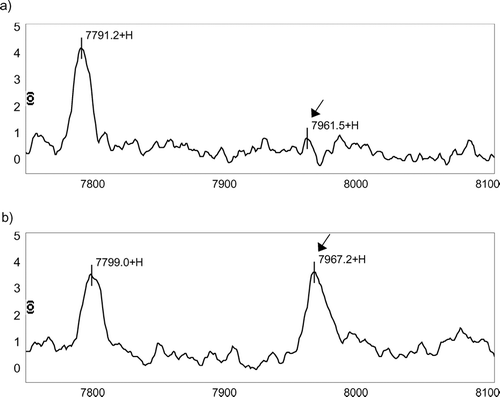
Figure 2 Best classification tree using IMAC F4 data. There are 5 peaks, which were used to construct the tree (8,170.82 Da, 17,397.9 Da, 7,961.19 Da, 6,975.67 Da, and 9,446.12 Da.). There are 7 nodes because 7,961.19 Da and 6,975.67 Da have double splits. Each node is sequentially labeled and shows splitting criteria. For instance, M8170_82 ≤ 1.586 would mean that subjects with peak intensities of ≤ 1.586 at m/z ratio at 8170.82 Da would move down the left side and all other subjects would move down the right side. A misclassification is defined when after “dropping” a case down a tree (i.e. following the classification rules of a tree), the case is misclassified as COPD when it is a control or control when it is COPD. Subjects continue down the tree until they reach terminal nodes. The number of samples that are at each node is given for both COPD and control (no COPD) groups. A terminal node is classified as COPD if the majority of samples are in the terminal node are from COPD patients. Otherwise the terminal node is classified as control (no COPD). Note that all subjects had protein chips run in duplicate so that the total number of samples is 120. Validation of sensitivity and specificity of the tree is done by “dropping” unknown samples down the tree and determining the percentage that are correctly classified.
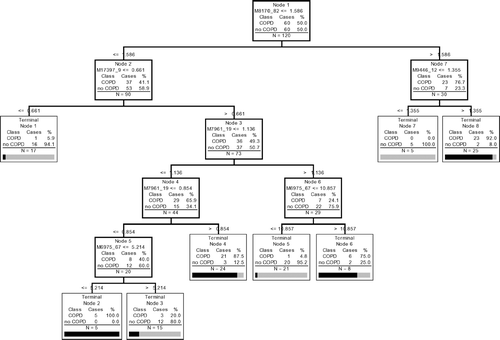
Figure 3 Minimum cost curves used to identify the number of nodes required to obtain a tree with the lowest cost. The x-axis represents trees with 1-11 terminal nodes and the y-axis shows relative cost assuming that there is an equal penalty for misclassifying both COPD subjects and controls.
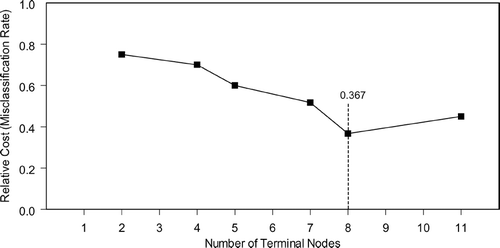
Figure 4 Receiver Operating Characteristic (ROC) curve of the tree shown in . The area under the ROC curve is 0.932.
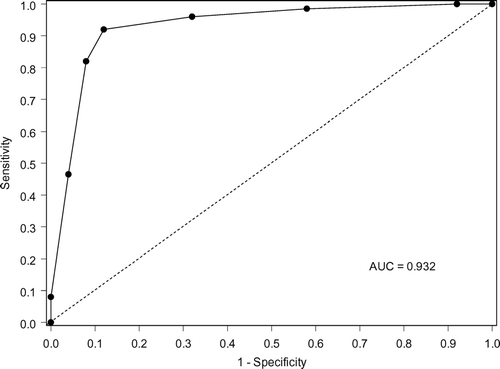
Figure 5 2-dimensional representation of the multidimensional data from CART shown in . (a) The root node, which is Node 1 in , contains all the subjects and their duplicates. They are split in two, left region (Node 2 in ) and right region (Node 7) with respect to the vertical line at intensity = 1.586 at 8170.82 Da. For the subjects in the left region, if their intensities at 17,397.9 are less than or equal to 0.661, they are classified as Terminal Node 1, no COPD. (b) If their intensities at 17,397.9 are greater than 0.661 (Node 3), they are further split in b with intensity levels at 7961.19 Da and 6975.67 Da. (c) The subjects on the right rectangle (Node 7) are further divided in two with the intensity level at 9,446.12 Da. A control subject is indicated by a cross (x) and a COPD subject indicated by a circle (o). (Continued)
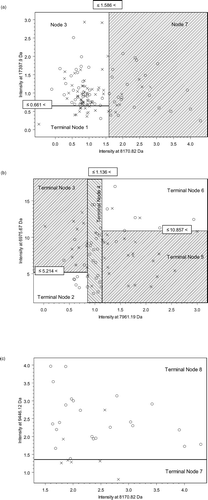
Table 3 Mean peak intensities of each member of the biomarker panel