Figures & data
Table 1a. The main characteristics and biomarkers in all COPD patient groups, all control groups and in sex-related groups.
Table 1b. The main characteristics and biomarkers in sex-related COPD and control groups according to smoking status.
Figure 1. Mean levels of urinary 8-oxodG/creatinine (nmol/mmol): a. in COPD and control groups according to smoking status; b. in COPD and control groups according to body mass index (BMI); c. in COPD group according to COPD stages. *difference between COPD and control groups (p < 0.05); #differences between former smokers with COPD group and smokers with COPD/non-smokers with COPD groups (p < 0.05); and differences between COPD with BMI < 18 group and COPD with BMI 18–25 group; §differences between very severe COPD with pack-year>30 group and severe COPD with pack-year > 30 group.
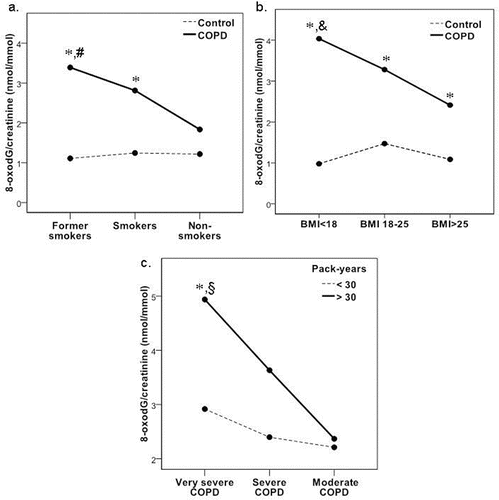
Table 2. Distribution of genotypes (dominant model) and gene combination of Xenobiotic-metabolising enzymes (XMEs) in all COPD patients, all control groups and in populations related to smoking status (smokers, non-smokers and former smokers).
Figure 2. Distribution of GSTM1/GSTT1 genotype combinations (with at least one deletion and without deletion): a. in all COPD and control groups; b. in non-smokers with COPD and control groups; c. in smokers with COPD and control groups; d. in former smokers with COPD and control groups.
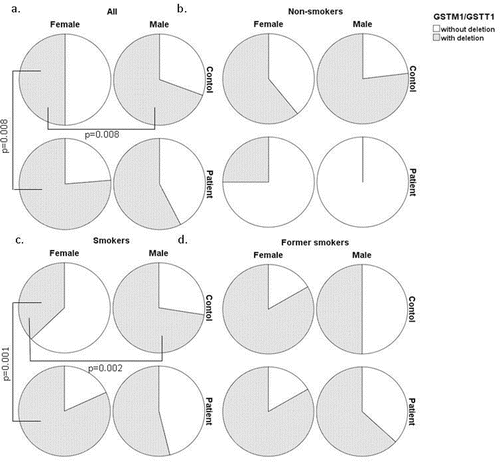
Table 3. Binary logistic regression model for predicting the COPD development in all investigated subjects and sex-related populations.
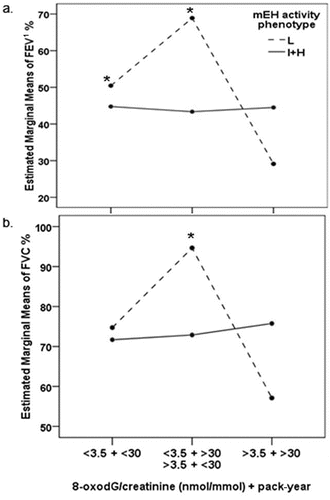
Figure 3. Combined effect of high oxidative stress (urinary 8-oxodG/creatinine > 3.5 nmol/mmol), heavy smoking (pack-year > 30) and mEH activity phenotype (L-low, I-intermediate and + H-high) on the pulmonary function assessed by: a. FEV1%; b. FVC%. * difference compared to the group of COPD patients with low mEH activity phenotype, high level of oxidative stress and heavy smoking habits (>3.5 + >30).