Figures & data
Table 1. Lung function examination in rats from each group.
Figure 1. Histopathologic changes of lung tissues from control group (A), emphysema group (B), and emphysema + SRT2104 group (C). Sections were stained with hematoxylin and eosin. Magnification, ×200.
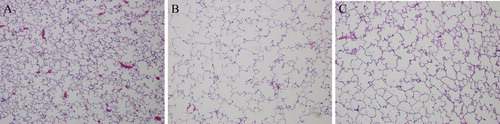
Table 2. Quantitative analyses of lung histomorphology in rats from each group.
Figure 2. SPA and SPC mRNA expression in lung tissues. All analyses were performed in triplicate. The relative expression of SPA and SPC mRNA was calculated with the 2−ΔΔCt method. Values are presented as mean ± SD (n = 15). *p < 0.05 vs. group A; #p < 0.05 vs. group B.
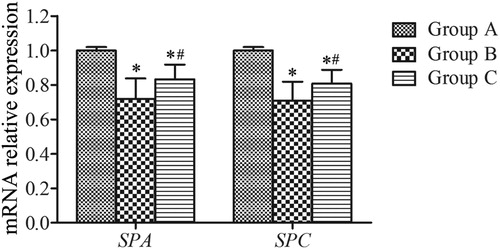
Figure 3. Western blotting analysis of the expression of SPA and SPC in lung tissues. β-actin was provided as the loading control. The expression of protein was analyzed by densitometry and normalized with β-actin. All analyses were performed in triplicate. Values are presented as mean ± SD (n = 15). *p < 0.05 vs. group A; #p < 0.05 vs. group B.
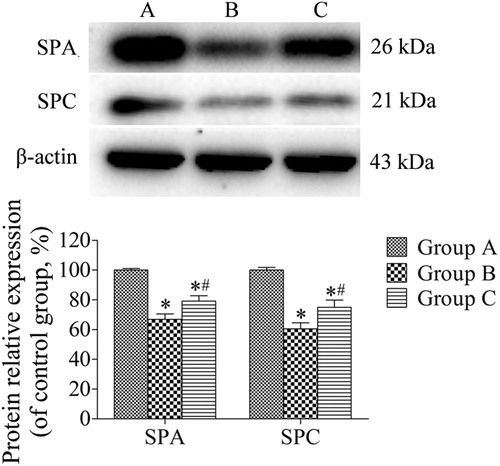
Figure 4. Immunohistochemical analysis of the expression of SPA in lung parenchyma from control group (A), emphysema group (B), and emphysema + SRT2104 group (C). The arrows indicate the SPA positive cells. Magnification, ×400. (D) The percentage of SPA positive cells in each group. Values are presented as mean ± SD (n = 15). *p < 0.05 vs. group A; #p < 0.05 vs. group B.
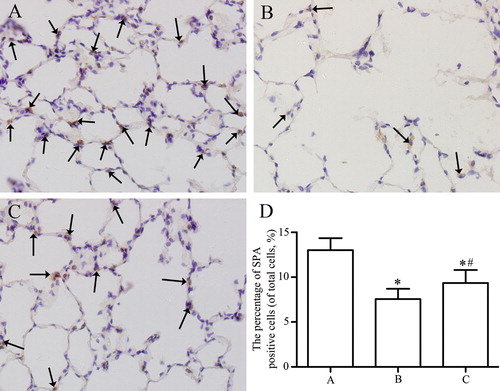
Figure 5. Immunohistochemical analysis of the expression of SPC in lung parenchyma from control group (A), emphysema group (B), and emphysema + SRT2104 group (C). The arrows indicate the SPC positive cells. Magnification, ×400. (D) The percentage of SPC positive cells in each group. Values are presented as mean ± SD (n = 15). *p < 0.05 vs. group A; #p < 0.05 vs. group B.
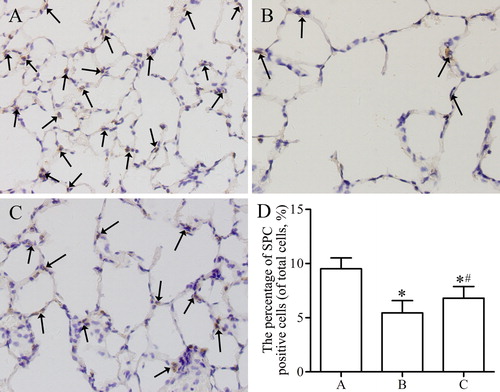
Figure 6. SA-β-gal activity of lung tissues from control group (A), emphysema group (B), and emphysema + SRT2104 group (C). The cells in lung samples with blue color were considered SA-β-gal positive, which means senescent cells. The arrows indicate the SA-β-gal-positive cells. Magnification, ×200.
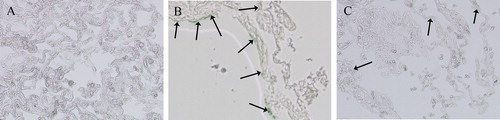
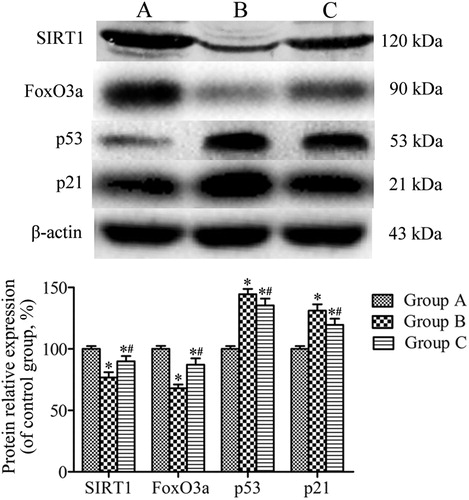
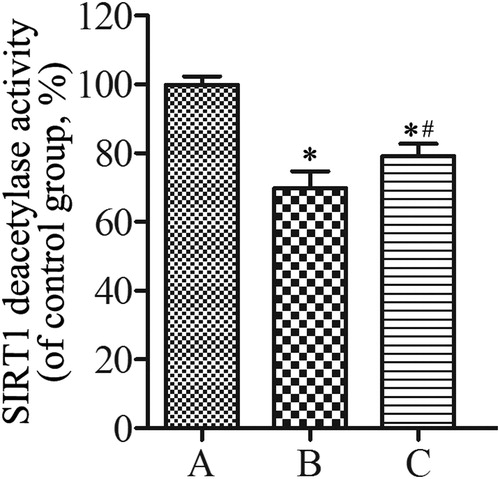
Figure 7. Western blotting analysis of the expression of SIRT1, FoxO3a, p53, and p21 in lung tissues. β-actin was provided as the loading control. The expression of protein was analyzed by densitometry and normalized with β-actin. All analyses were performed in triplicate. Values are presented as mean ± SD (n = 15). *p < 0.05 vs. group A; #p < 0.05 vs. group B.