Figures & data
Figure 1. Directed acyclic graphs depicting three possible scenarios that could explain cause effects between interleukin (IL)-1/IL-1 receptors/receptor antagonists (IL-1Rs) and airflow obstruction (AO). (A) IL-1/IL-1Rs have effects on AO. (B) AO has reverse effects on IL-1/IL-1Rs. (C) IL-1 has an indirect effect on AO, perhaps via its receptors (IL-1Rs). Under the multivariable MR framework, the direct effect of IL-1 on AO is estimated as β, whilst its indirect effect is estimated as αγ. The total effect can be estimated as β+αγ .
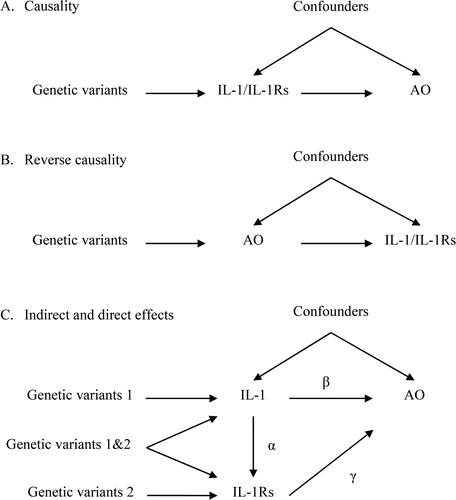
Figure 2. The analytical framework of two-sample Mendelian randomization (MR) used in this study. Cis-pQTLs: cis protein quantitative trait loci; COPD: chronic obstructive pulmonary disease; ICGC: International COPD Genetics Consortium.
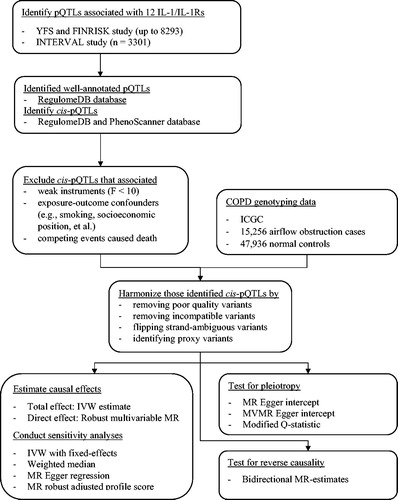
Figure 3. Causal estimates of genetically predicted interleukin (IL)-1α/β, IL-1 receptor antagonist (IL-1Ra), IL-1 recetpor-type 1 (IL-R1), and IL-1 receptor accessory protein (IL-1Racp) with airflow obstruction (AO) and the negative control outcome of tuberculosis using univariable and multivariable Mendelian randomization analyses. Multivariable MR* adjusted either IL-1
α/β or IL-1Ra; and multivariable MR** adjusted either IL-1
α/β, IL-1Ra, IL-1R1, IL-1Racp, IL-18, IL-18BP, IL-18Rα
, IL-36α/β/γ
, or IL-37. The modified Q-statistic for multivariable MR* estimates was 6.63 with p = 0.675, and the corresponding value for multivariable MR** estimates was 33.48 with p = 0.945. IL-18: interleukin-18; IL-18BP: interleukin-18 binding protein; IL-18Rα: interleukin-18 receptor 1; IL-36α/β/γ: interleukin-36 α/β/γ; IL-37: interleukin-37.
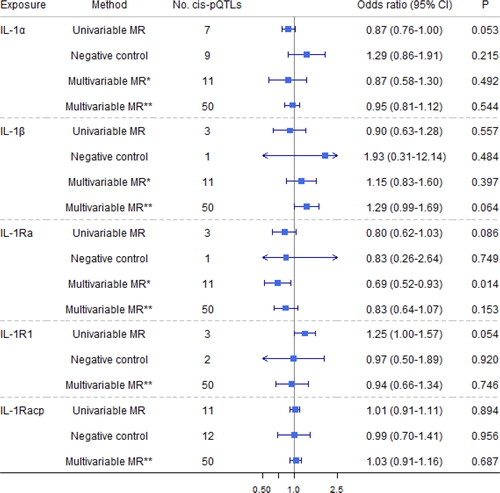
Figure 4. Causal estimates of genetically predicted interleukin (IL)-18, IL-18 binding protein (IL-18BP), IL-18 receptor 1 (IL-18Rα
), and IL-37 with airflow obstruction (AO) and the negative control outcome of tuberculosis using univariable and multivariable Mendelian randomization analyses. Multivariable MR* adjusted either IL-18, IL-18BP, IL-18Rα
, or IL-37; and multivariable MR** adjusted either IL-1
α/β, IL-1Ra, IL-1R1, IL-1Racp, IL-18, IL-18BP, IL-18Rα
, IL-36α/β/γ
, or IL-37. The modified Q-statistic for multivariable MR* estimates was 4.36 with p = 0.824, and the corresponding value for multivariable MR** estimates was 33.48 with p = 0.945. IL-1α: interleukin-1 α; IL-1β: interleukin-1 β; IL-1Ra: interleukin-1 receptor antagonist; IL-1R1: interleukin-1 receptor-type 1; IL-37: interleukin-37.
Figure 5. Causal estimates of genetically predicted interleukin (IL)-36α, IL-36β
, and IL-36γ
with airflow obstruction (AO) and the negative control outcome of tuberculosis using univariable and multivariable Mendelian randomization analyses. Multivariable MR* adjusted either IL-36α
, IL-36β
, or IL-36γ
; and multivariable MR** adjusted either IL-1
α/β, IL-1Ra, IL-1R1, IL-1Racp, IL-18, IL-18BP, IL-18Rα
, IL-36
α/β/γ, or IL-37. The modified Q-statistic for multivariable MR* estimates was 1.52 with p = 0.997, and the corresponding value for multivariable MR** estimates was 33.48 with p = 0.945. IL-1α: interleukin-1 α; IL-1β: interleukin-1 β; IL-1Ra: interleukin-1 receptor antagonist; IL-18: interleukin-18; IL-18BP: interleukin-18 binding protein; IL-1R1: interleukin-1 receptor-type 1; IL-18Rα: interleukin-18 receptor 1; IL-37: interleukin-37.
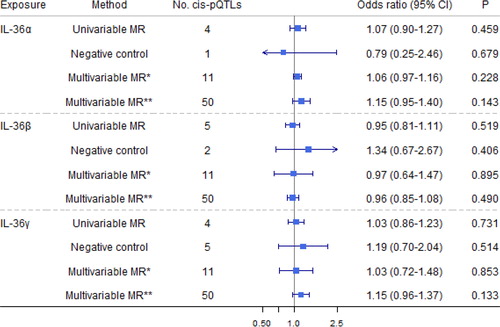
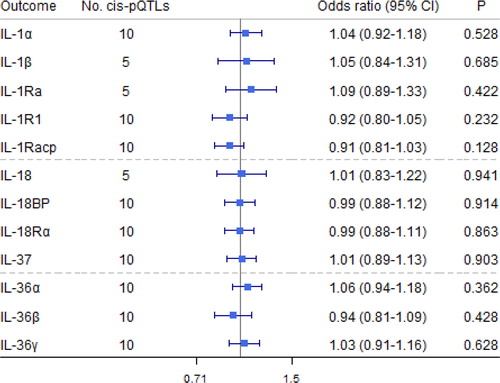
Figure 6. Causal estimates of genetically predicted airflow obstruction (AO) on interleukin (IL)-1 and their receptors and receptor antagonists using univariable Mendelian randomization analyses. IL-1α: interleukin-1 α; IL-1β: interleukin-1 β; IL-1Ra: interleukin-1 receptor antagonist; IL-18: interleukin-18; IL-18BP: interleukin-18 binding protein; IL-1R1: interleukin-1 receptor-type 1; IL-18Rα: interleukin-18 receptor 1; IL-36α/β/γ: interleukin-36 α/β/γ; IL-37: interleukin-37.
Data availability
The summary statistics for genetic associations of IL-1α, IL-1R1, IL-1Racp, IL18BP, IL18Rα, IL-36α, IL-36β, IL-36γ, IL-37 are available in the INTERVAL study (http://www.phpc.cam.ac.uk/ceu/proteins/). The summary statistics for genetic associations of IL-1β, IL-1Ra, IL-18 are available in YFS and FINRISK survey (https://grasp.nhlbi.nih.gov/FullResults.aspx). The summary genetic statistics for genetic associations of chronic obstructive pulmonary disease are available in the Genetic Epidemiology of COPD (dbGaP study accession: phs000179.v6.p2). The annotated QTLs are available in RegulomeDB (https://regulomedb.org/regulome-search/) and PhenoScanner (http://www.phenoscanner.medschl.cam.ac.uk/). The 1000 genomes European reference panel is available via LDlink (https://ldlink.nci.nih.gov/).
Code availability
Software codes and data are available on request.
