Figures & data
Table 1. Demographic data used to assess STK11 protein levels.
Table 2. Demographic data used to assess STK11 mRNA levels.
Table 3. List of antibodies used.
Table 4. List of primers used.
Figure 1. Smokers with COPD display diminished pulmonary STK11 protein expression. (A) Immunoblot analysis showing the STK11 protein levels in the lung tissues of smokers with normal lung function (control) and COPD. (B) The STK11 densitometry data (normalized to Hypoxanthine Phosphoribosyltransferase 1 [HPRT1]) derived from (A). (C) Expression of STK11 mRNA from the lung tissues of smokers with normal lung function (control) and COPD. The expression was determined by RT-PCR, and the change in expression relative to GAPDH (control gene) was presented. Data are represented as median with IQR and analyzed using by student’s t-test.
![Figure 1. Smokers with COPD display diminished pulmonary STK11 protein expression. (A) Immunoblot analysis showing the STK11 protein levels in the lung tissues of smokers with normal lung function (control) and COPD. (B) The STK11 densitometry data (normalized to Hypoxanthine Phosphoribosyltransferase 1 [HPRT1]) derived from (A). (C) Expression of STK11 mRNA from the lung tissues of smokers with normal lung function (control) and COPD. The expression was determined by RT-PCR, and the change in expression relative to GAPDH (control gene) was presented. Data are represented as median with IQR and analyzed using by student’s t-test.](/cms/asset/87b543c3-184b-41c0-95dd-d677e9aed349/icop_a_2342797_f0001_b.jpg)
Figure 2. STK11 protein expression is decreased in CS-exposed mice. Mice were exposed to CS or RA for four days. (A) Whole-lung tissue protein lysates were prepared, and the STK11 protein expression was determined by immunoblotting. (B) The STK11 densitometry data normalized to β-Actin. (C) The STK11 densitometry data normalized to HPRT1. (D) Total RNA from whole lung tissue was prepared, and the expression levels of STK11 mRNA were determined by RT-PCR. The change in expression relative to β-Actin (control gene) was presented. Data are represented as median with IQR and analyzed using by student’s t-test.
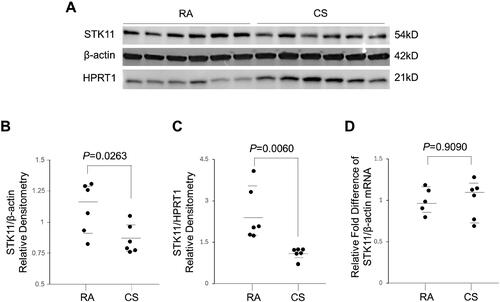
Figure 3. STK11 attenuates CS-induced BEAS-2B cell cytotoxicity. BEAS-2B cells were cultured in the presence and absence of CSE, as indicated above. Whole-cell proteins and Total RNA were isolated, and STK11 expression was determined by immunoblotting (A and B) and RT-PCR (C–F). The mRNA expression levels were measured in triplicate using the same BEAS-2B cells. The bar graph indicates mean ± SE. STK11 was overexpressed in cells by transfecting with STK11-FLAG tag plasmid for 48 h. (G) Whole-cell proteins were isolated and immunoblotted with anti-FLAG (STK11) or anti-β-actin antibodies to detect the STK11 overexpression. (H) MTT assay was performed to measure the cell viability following transfection and culture in the presence or absence of CSE (2%) for 24 h. BEAS-2B cells were transfected with scrambled siRNA (siCtrl) or STK11-targeted siRNAs for 72 h to knockdown STK11. (I) Whole-cell proteins were isolated and immunoblotted with STK11 or β-actin antibodies to detect the STK11 knockdown. (J) MTT assay was performed to measure the cell viability, following siSTK11#3 transfection and culture in the presence or absence of 2% of CSE (2%) for 24 h. All experiments were repeated three times. Data are expressed as mean ± SEM and analyzed using one-way ANOVA followed by a Bonferroni’s multiple-comparison correction test.
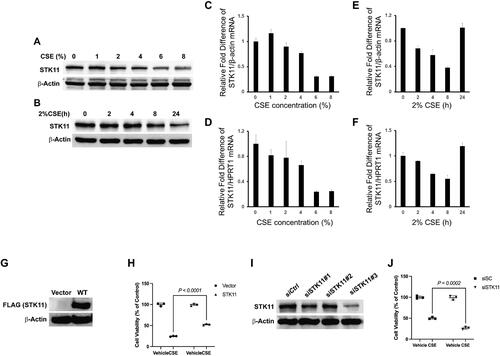
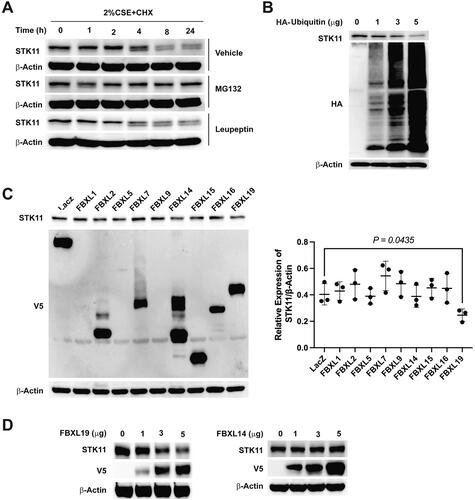
Figure 4. FBXL19 targets STK11 for proteasomal degradation. (A) To determine STK11 stability in BEAS-2B cells, cells were treated with 20 μg/mL of cycloheximide in the presence of 40 μM of MG132 or 20 μM of leupeptin indicated in the figure. Whole-cell proteins were isolated and immunoblotted with STK11 or β-actin antibodies. (B): Cells were transfected with ubiquitin-HA plasmids as indicated, whole cell proteins were isolated, and the expression of STK11, HA, or β-actin was determined by immunoblotting. To find the E3 ligase that explicitly recognizes and facilitates STK11 degradation, (C) FBXL–V5-tagged family plasmids were transfected in cells, and whole-cell proteins were immunoblotted with anti-STK11, anti-V5 or anti-β-actin antibodies. (D) BEAS-2B cells were transfected as indicated with FBXL19 or FBXL14 –V5 tagged plasmids, and whole-cell proteins were immunoblotted with anti-STK11, anti-V5, or anti-β-actin antibodies. All experiments were repeated three times.
Data availability statement
The datasets used and analyzed in this study are available from the corresponding author upon reasonable request.