Figures & data
TABLE 1 Overview of Rattus norvegicus connexins detected and sequenced, and their accession numbers in GenBank
TABLE 2 Overview of Mesocricetus auratus connexins and accession numbers in GenBank
TABLE 3 Overview of Cricetulus griseus connexins and their accession numbers in GenBank
TABLE 4 Primer-combinations for screening of connexin expression in Syrian hamster (Mesocricetus auratus) organs and cells, V79 cells (Chinese hamster, Cricetulus griseus) and mouse (Mus musculus, Mm) organs
TABLE 5 Expression of connexins in some Syrian hamster organs, SHE cells and V79 cells as detected by RT-PCR
Figure 1 Similarities and differences in connexin expression in hamster and mouse. Expression of (A) Cx29, (B) Cx39, (C) Cx46, and (D) Cx57 in some hamster organs and cells (left panels) and in some mouse organs (right panels). RNA was isolated from several Syrian hamster and mouse organs, Syrian hamster embryo (SHE) cells, and Chinese hamster V79 cells. The RNA was DNase treated. After removal of DNase, the samples were divided in two. One part was reverse transcribed, the other part was handled similarly but without reverse transcriptase. PCR was then performed on both samples. The upper panels show samples that were reverse transcribed, and the lower panels show the corresponding samples that were not reverse transcribed, indicating the lack of genomic contamination. The positive controls (pos.) are samples that have not been cleaned from genomic contamination. The positions of size markers are indicated to the left.
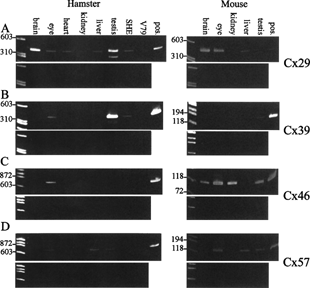
TABLE 6 Connexin expression profiles in wild-type and Min mouse as detected by RT-PCR