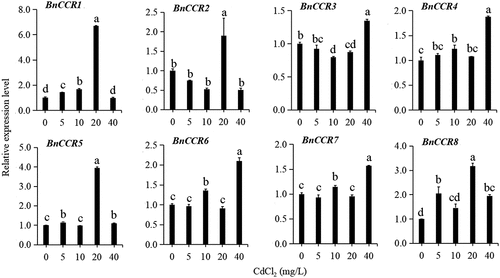
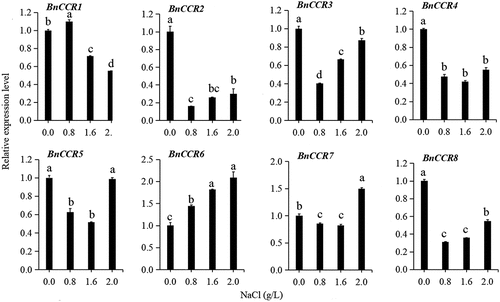
Figures & data
Figure 1. Schematic diagram CCR reaction process in lignin biosynthesis of ramie.
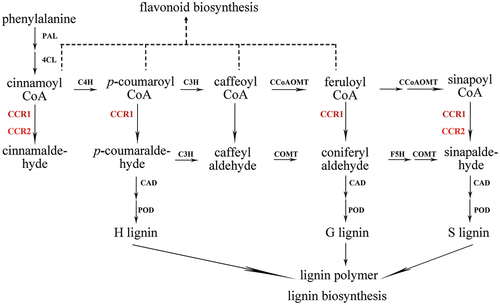
Figure 2. Gene structure of BnCCR, CDS, UTR, and introns are represented by black rectangle, gray rectangles, and gray lines, respectively (a). Phylogenetic relationship of the CCR proteins of ramie and other plants (b).
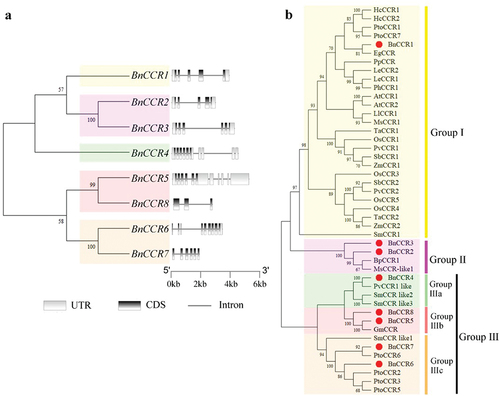
Table 1. Analysis of the conserved and active sites of BnCCR proteins.
Figure 3. Structural modeling analysis of BnCCR proteins. The extended strand is indicated by a yellow ribbon, the alpha helix is indicated by a pink ribbon, the beta-turn and the random coil is indicated by a white ribbon, the R-(X)5-K motif is indicated by red stick amino acid residues, the conserved sites S-Y-K of the short chain dehydrogenase superfamily is indicated by blue, earthy yellow, and green stick amino acid residues, respectively. The important substrate binding site H202 of CCR is indicated by a green round ball, and NAP in BnCCR1 is shown in light blue rod amino acid residues.
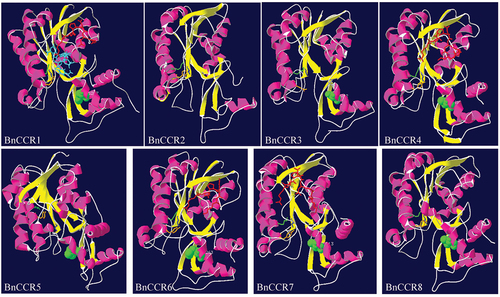
Figure 6. The expression profile of BnCCR genes in the xylem and phloem of the stem at the two periods (a), the lignin content profile of the xylem and phloem of the stem at the two periods (b).
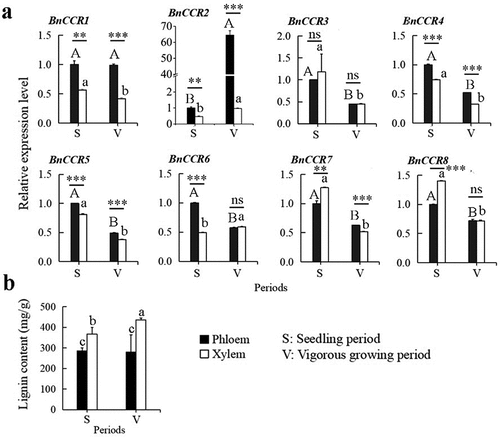
Table 2. Correlation coefficients of lignin content and the expression levels of BnCCR genes.
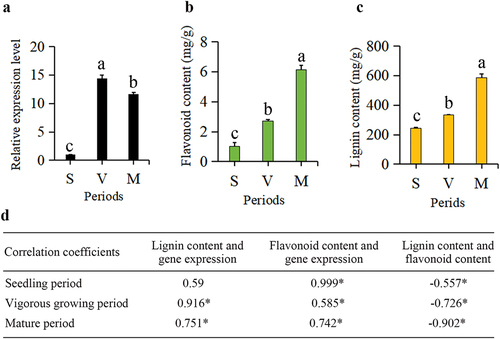
Figure 7. The expression profile of BnCCR2 in ramie during three periods (a), histogram of the flavonoid content in ramie during three periods (b), histogram of the lignin content in ramie during three periods (c), correlation coefficients of the lignin content, flavonoid content and the expression level of BnCCR2 (d). S: seedling period, V: vigorous growing period, M: mature period.