Figures & data
TABLE 1. Oligonucleotide primers for real-time PCR.
FIGURE 1. Characterization of hepatocytes and hUCB-MSCs and Fabrication of 3D printed alginate structure. (A). (a) Confirmation of hepatocyte morphology, (b) fluorescence expression in CAG-dsRed primary hepatocytes, (c) Alb (green), AFP(red), (d) CK18 (green), and CYP1A2 (red). (e) Morphology of hUCB-MSCs, (f) GFP-tagged, and (g and f) confirmation of differentiation ability into adipogenic and osteogenic linages of hMSCs. Bars, 50 μm (c-d), 100 μm (a-b, e-f), and 200 μm (g-h). (B). A schematic diagram of a 3D printed architecture composed of mouse primary hepatocytes and human MSCs. The 3D hepatic architecture manufactured to be 25 mm x 25 mm in size and composed of 5 layers. (C). Distribution of CAG-dsRed primary hepatocytes and GFP-tagged hMSCs in the 3D hepatic architecture under fluorescence microscopy at day 1. Bars, 100 μm.
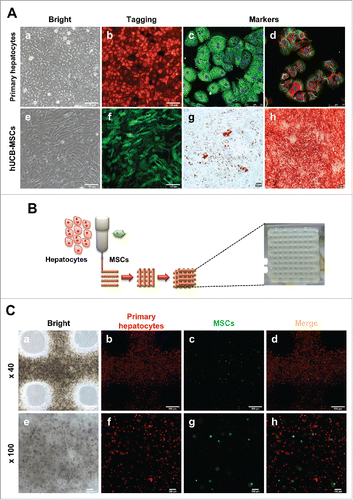
FIGURE 2. Comparison of morphology between 2D culture dish and 3D alginate architecture. (A). ∼ (C) Morphological changes onto 2D culture dish. Cuboidal hepatic morphology is forfeited as time goes on until day 7. (D)∼(F) Morphological changes compared between 2D. Cuboidal hepatic morphology is sustained until day 7 in 3D alginate architecture. (G) Magnification of (F). Black arrows indicate areas mimicking hepatic structure in 3D architectures. Bars, 100 μm.
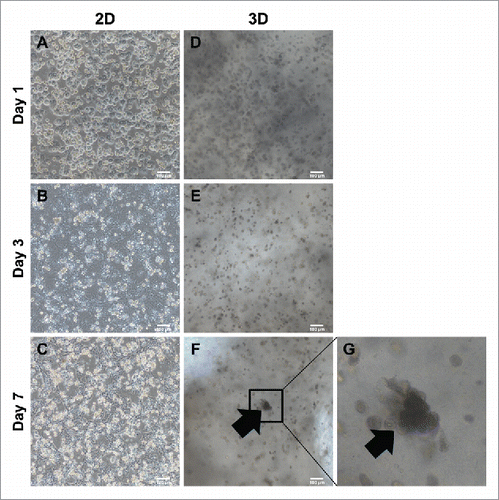
FIGURE 3. Migration and colonization of mouse primary hepatocytes with hMSCs. (A) and (E) Bright microscope image of 3D printed mouse primary hepatocytes and hMSCs. (B) ∼ (D) and (F) ∼ (H) Fluorescence image of 3D printed mouse primary hepatocytes and hMSCs. Bars, (A) ∼ (D) is 500 μm and (E) ∼ (F) is 100 μm.
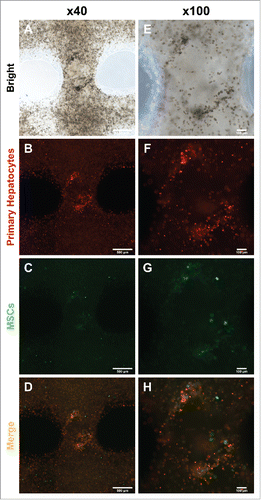
FIGURE 4. Comparison of hepatic functions between the 2D culture dish and 3D alginate architecture. (A). The expression of Albumin, ASGR1, CK18 and CYP1A2 genes gradually increased in primary hepatocytes cultured with hMSCs in a 3D hepatic structure. Gene expression continued for at least 7 days. MEF, mouse embryonic fibroblasts. *P < 0.05, **P < 0.01, ***P < 0.001. Data are mean ± SD. (B). Mouse Albumin secretion was measured in the cultured medium at day 1, 3, and 7. *P < 0.05, **P < 0.01. Data are mean ± SD. (C). Urea synthesis level at day 7 in hepatocytes cultured in 2D, 3D and 3D with MSCs. **P < 0.01, ***P < 0.001. Data are mean ± SD.
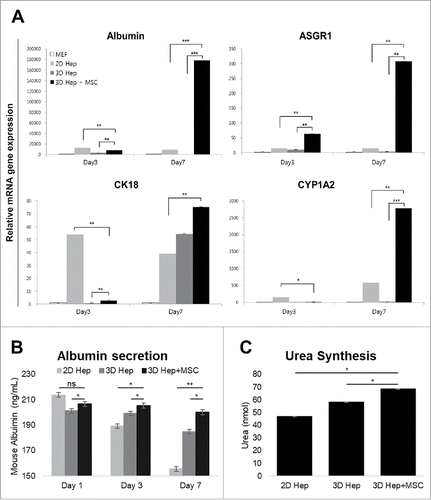
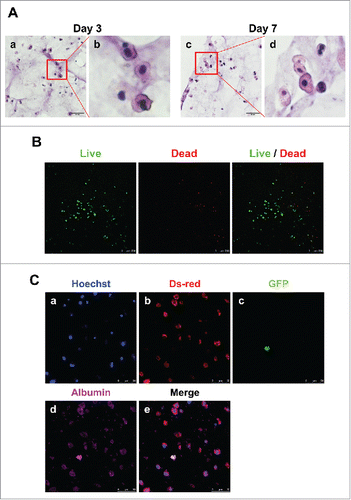
FIGURE 5. Detection of hepatocytes and MSCs in 3D architecture by H&E staining, live/dead assay and immunostaining. (A). H&E staining at day 3 (a, enlarged in b) and 7 (c, enlarged in d). Bars, 60 μm. (B). Live/Dead Viability/Cytotoxicity Assay. Live cells (Green) and Dead cells (Red). Bars, 50 μm. (C). Immunofluorescent staining for DsRed (b), GFP (c) and albumin (d) at day 7. Hoechst (blue; a) labels all nuclei. Bars, 50 μm.