Figures & data
FIGURE 1. LRRC 19 expression is induced during I/R injury. A) The average size of the wound area in mice (n = 10) after I/R injury were measured at day 0, 1, 3, 7 10 and 14 post injury. The end of 2nd ischemia was assigned day 0. The size of the ulcer on day 3 was assigned a value of 100%. B) Realtime PCR analysis of LRRC19 mRNA expression in the I/R site. The end of 2nd ischemia was assigned day 0. Data are relative to mRNA level on day 0. Values were determined in n = 10 mice. C) Western blot analysis of LRRC19 protein expression at the wound site during I/R injury for the indicated time points. The end of 2nd ischemia was assigned day 0. The data are representative of n = 10 subjects. The band intensity of LRCC19 was normalized against loading control. The normalized band densitometry at day 0 was set as baseline of value 1.
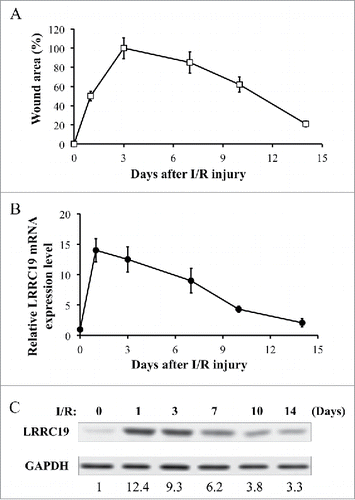
FIGURE 2. Knocking down LRRC19 reduces pressure ulcer formation and decreases the inflammatory cell infiltration in wound area. A) Western blot analysis of LRRC19 protein expression at the wound site during I/R injury. The wound area was topically applied with 100 pmol of either LRRC19 siRNA or control siRNA immediately after 1st ischemia delivery, followed by day 0, 1, 3, 6, 9, 12 after I/R injury induced. The end of 2nd ischemia was assigned day 0. The data are representative of n = 4 subjects for each group. The band intensity of LRCC19 was normalized against GAPDH. The normalized band densitometry of the siRNA control treated sample at day 0 was set as baseline of value 1. B) The average size of the wound area in mice (n = 6 for each group) treated with either LRRC19 or control siRNA during I/R injury were measured at day 0, 1, 3, 7 10 and 14 post injury. The end of 2nd ischemia was assigned day 0. The size of the ulcer in control group on day 3 after I/R was assigned a value of 100%. Immunohisochemistry was used to study the number of C) neutrophils (myeloperoxidase positive) and D) macrophages (CD68 positive) at the lesion site. Values were determined in six random microscopic fields (∼20,000 mm2) in n = 6 mice per group. The results are presented as cells per mm2. The data are presented as the mean ± SD (n = 4 for each time point). *p < 0.05, **p < 0.01, significantly different from control group.
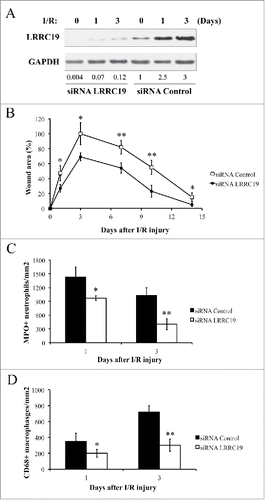
FIGURE 3. Knocking down LRRC19 suppresses proinflammatory cytokines production during I/R injury. ELISA analysis of A) TNFα, B) IL-6 and C) IL-1β at the wound site during I/R injury at the indicated time points. The wound area was topically applied with 100 pmol of either LRRC19 siRNA or control siRNA immediately after 1st ischemia delivery, followed by day 0, 1 after I/R injury induced. The end of 2nd ischemia was assigned day 0. The data are presented as the mean ± SD (n = 4). *p < 0.05, **p < 0.01, significantly different from control group measured on the same day.
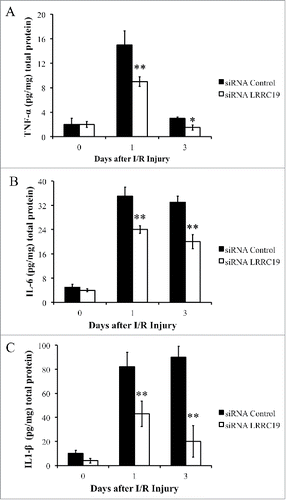
FIGURE 4. Knocking down LRRC19 suppresses NFκB signaling. Western blot analysis of NFκB signaling components. Primary dermal fibroblasts transfected with either scramble siRNA or LRRC19 siRNA were challenged with 50 ng/ml IL-1β for the indicated time points. Cell lysate after treatment was subjected to SDS-PAGE to determine the level of A) total p65, phospho-p65 (S536), total IkBα and phosphor-IkBα (S32); or the level of B) COX-2 and iNOS. The band intensity of each target protein was normalized against its corresponding loading control. The normalized band densitometry of the siRNA control transfected sample at day 0 was set as baseline of value 1. The data are representative of at least three independent experiments.
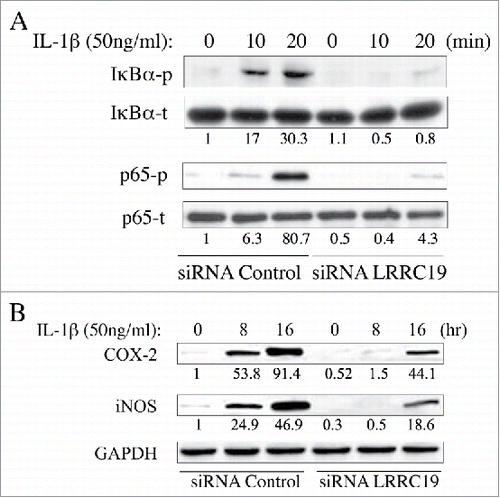
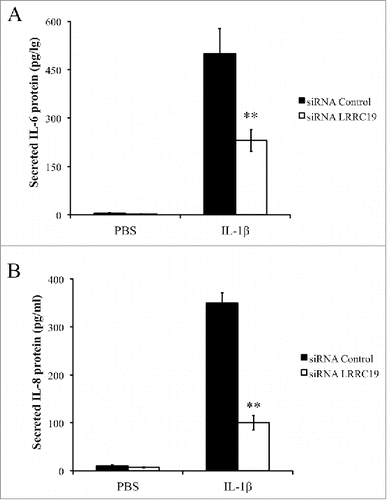
FIGURE 5. Knocking down LRRC19 reduces IL-1β mediated proinflammatory cytokines production. Primary dermal fibroblasts transfected with either scramble siRNA or LRRC19 siRNA were challenged with 50 ng/ml IL-1β or PBS for 24 h. The cell culture supernatant was then harvested to determine the concentrations of A) IL-6 and B) IL-8 by ELISA. The data are presented as the mean ± SD (n = 4). *p < 0.05, **p < 0.01, significantly different from PBS control group.