Figures & data
Table 1. Primers nucleotide sequence
Figure 1. Histological features of hepatocyte differentiation in decellularized liver scaffold (Mag. 400x). A: HE-stained Liver control, B: HE-stained decellularized liver scaffold, C: HE-stained hepatocyte-differentiated MSC d7 in decellularized liver scaffold, D: HE-stained hepatocyte- differentiated iPSC d7 in decellularized liver scaffold, E: HE-stained hepatocyte-differentiated MSC d21 in decellularized liver scaffold, F: HE-stained hepatocyte-differentiated iPSC d21 in decellularized liver scaffold, G: Masson trichrome-stained liver control, H: Masson trichrome-stained decellularized liver scaffold, I: Masson trichrome-stained hepatocyte-differentiated MSC d7 in decellularized liver scaffold, J: Masson trichrome-stained hepatocyte-differentiated iPSC d7 in decellularized liver scaffold, K: Masson trichrome-stained hepatocyte-differentiated MSC d21 in decellularized liver scaffold, L: Masson trichrome-stained hepatocyte-differentiated iPSC d21 in decellularized liver scaffold cells suspected of being MSCs were clearly observed (marked with circles), M: Brightfield image from inverted microscope of hepatocyte-differentiated iPSC d7 in decellularized liver scaffold, N: Brightfield image from inverted microscope of hepatocyte-differentiated iPSC d7 in decellularized liver scaffold. Circles marked cell adherence to scaffold. O. Quantification collagen area using Image J measurement from hepatocyte differentiated MSCs (n = 3) or iPSCs (n = 3) in decellularized liver scaffold (%).
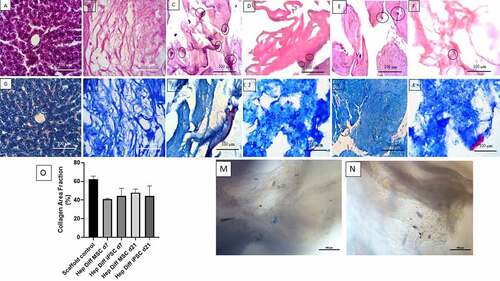
Figure 2. Representative SEM images of hepatocyte differentiation in decellularized liver scaffold (Mag. 5000x). A: hepatocyte-differentiated iPSC d7 in decellularized liver scaffold (n = 3), B: hepatocyte-differentiated iPSC 21 in decellularized liver scaffold (n = 3), C: hepatocyte-differentiated MSC d7 in decellularized liver scaffold (n = 3), F: hepatocyte-differentiated MSC d21 in decellularized liver scaffold (n = 3). Circles marked cell adherence to scaffold.
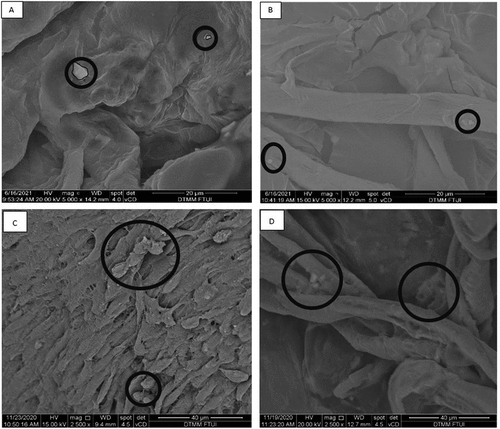
Figure 3. Albumin detection from hepatocyte differentiation in decellularized liver scaffold. A: IHC albumin liver negative control (Mag. 400x), B: IHC albumin liver positive control (Mag. 400x), C: IHC albumin hepatocyte-differentiated MSC d7 in decellularized liver scaffold (Mag. 40x), D: IHC albumin hepatocyte-differentiated iPSC d7 in decellularized liver scaffold (Mag. 40x), E: IHC albumin hepatocyte-differentiated MSC d21 in decellularized liver scaffold (Mag. 40x), F: IHC albumin hepatocyte-differentiated iPSC d21 in decellularized liver scaffold (Mag. 40x), G: Percentage of albumin area fraction using Image J measurement from hepatocyte-differentiated MSCs in decellularized liver scaffold (n = 3), H: Percentage of albumin area fraction using Image J measurement from hepatocyte-differentiated iPSCs in decellularized liver scaffold (n = 3), I: qRT-PCR mean normalized albumin gene expression from hepatocyte-differentiated MSCs in decellularized liver scaffold (n = 3), J: qRT-PCR mean normalized albumin gene expression from hepatocyte-differentiated iPSCs in decellularized liver scaffold (n = 3). Comparison between groups is not significant (p > .5).
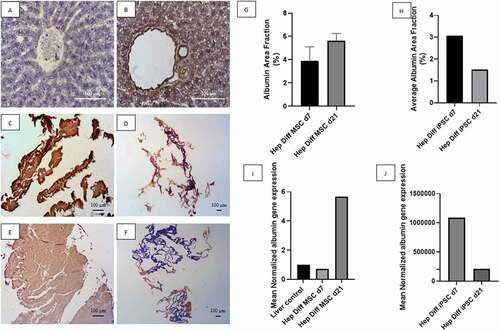
Figure 4. CYP450 identification from hepatocyte differentiation in decellularized liver scaffold (Mag. 400x). A: IHC CYP450 liver negative control, B: IHC CYP450 liver positive control, C: IHC CYP450 hepatocyte-differentiated MSC d7 in decellularized liver scaffold, D: IHC CYP450 hepatocyte-differentiated iPSC d7 in decellularized liver scaffold, E: IHC CYP450 hepatocyte-differentiated MSC d21 in decellularized liver scaffold, F: IHC CYP450 hepatocyte-differentiated iPSC d21 in decellularized liver scaffold, G: Percentage of CYP450 area fraction using Image J measurement from hepatocyte-differentiated MSCs in decellularized liver scaffold (n = 3), H: Percentage of CYP450 area fraction using Image J measurement from hepatocyte-differentiated iPSCs in decellularized liver scaffold (n = 3), I: qRT-PCR mean normalized CYP450 gene expression from hepatocyte-differentiated MSCs in decellularized liver scaffold (n = 3), J: qRT-PCR mean normalized CYP450 gene expression from hepatocyte-differentiated iPSCs in decellularized liver scaffold (n = 3). Comparison between groups is not significant (p > .5).
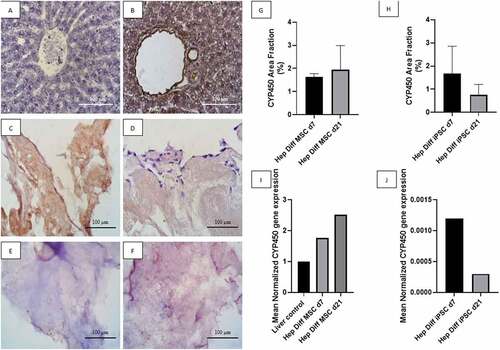
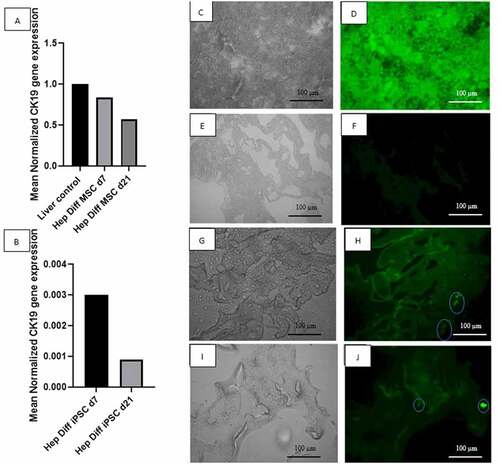
Figure 5. Determination of hepatocyte maturation stage by qRT-PCR CK-19 and IF HNF4-a and CEBPA. A: qRT-PCR mean normalized CK-19 gene expression from hepatocyte-differentiated MSCs in decellularized liver scaffold (n = 3), B: qRT-PCR mean normalized CK-19 gene expression from hepatocyte-differentiated iPSCs in decellularized liver scaffold (n = 3), C: Brightfield image from fluorescence microscope liver control, D: IF HNF4a liver-positive control, E: Brightfield image from fluorescence microscope decellularized liver scaffold, F: IF HNF4a decellularized liver scaffold control, G: Brightfield image from fluorescence microscope hepatocyte-differentiated iPSC d21 in decellularized liver scaffold, H: IF HNF4a hepatocyte-differentiated iPSC d21 in decellularized liver scaffold, I: Brightfield image from fluorescence microscope hepatocyte-differentiated iPSC d21 in decellularized liver scaffold, J: IF CEBPA hepatocyte-differentiated iPSC d21 in decellularized liver scaffold.