Figures & data
Table 1. Eukaryotic tRNA modifications that require 2-subunit enzymes.
Figure 1. S. cerevisiae tRNA modifications formed by 2-subunit enzymes are located throughout the tRNA. Cloverleaf schematic depicting tRNA residues known to be modified (black) in S. cerevisiae, and those that are not modified (gray). Modifications (in brackets) formed by 2-subunit enzymes are labeled.
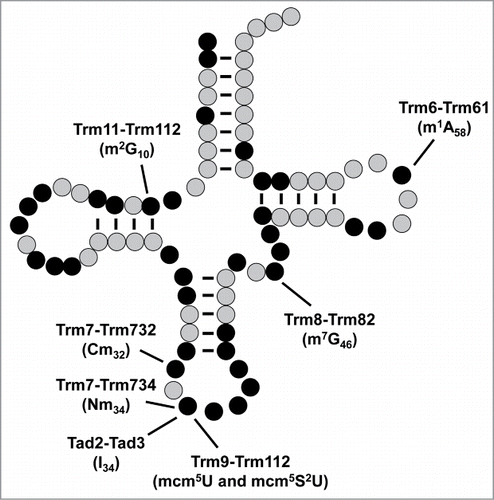
Figure 2. Schematic of FtsJ methyltransferase stem-loop substrates. S. cerevisiae Trm7 requires Trm732 for Cm32 modification and Trm734 for Gm34 modification of the anticodon loop of tRNAPhe which, as for all tRNAs, has 7 bases. These modifications then drive formation of yW37 from its m1G precursor. The thicker arrow from Gm34 indicates that yW formation is more dependent on this modification. Other FtsJ family members modify the 5-base A-loop in the rRNA large subunit in different organisms and organelles, as indicated.
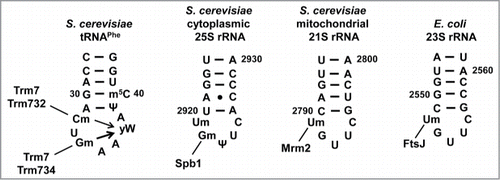
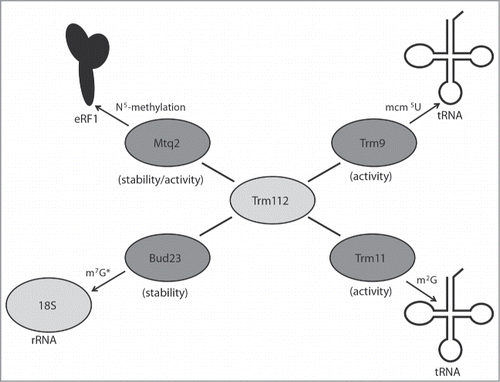
Figure 3. Trm112 partners with several methyltransferases involved in diverse translational processes. Schematic of the S. cerevisiae Trm112 physical interactions known to affect the activity or stability (in brackets) of its partner methyltransferases. (*) The Sc Bud23 protein, but not its methyltransferase activity, is required for m7G1575 formation on the cytoplasmic 18S rRNA subunit.