Figures & data
Figure 1. Schematic drawing for Tryptophan operon leader. The Trp codon (W) tandem is highlighted in green color in both the gene structure (the leader peptide sequence is shown) and the RNA secondary structures. The stem of the termination hairpin is colored in red in both terminator and anti-terminator structures. In the anti-terminator structure, the ribosome, stalling on the Trp tandem, impedes the formation of the first hairpin. The 5' portion (red) of the termination hairpin is then sequestered in an anti-terminator hairpin. The RNA secondary structures in are drawn via VARNA[Citation98].
![Figure 1. Schematic drawing for Tryptophan operon leader. The Trp codon (W) tandem is highlighted in green color in both the gene structure (the leader peptide sequence is shown) and the RNA secondary structures. The stem of the termination hairpin is colored in red in both terminator and anti-terminator structures. In the anti-terminator structure, the ribosome, stalling on the Trp tandem, impedes the formation of the first hairpin. The 5' portion (red) of the termination hairpin is then sequestered in an anti-terminator hairpin. The RNA secondary structures in Figures 1–4 are drawn via VARNA[Citation98].](/cms/asset/e57297a8-d7d7-4dd0-af4c-f901318e3453/krnb_a_1008373_f0001_oc.gif)
Figure 2. Schematic drawing for Levivirus (MS2). The UCS is highlighted in red color, the SD sequence is in yellow color, and the start codon is colored in green. In the left structure, the SD sequence is accessible to ribosome for translation. In the right structure, the SD pairs with the UCS, forming the LDI to impede ribosome binding.
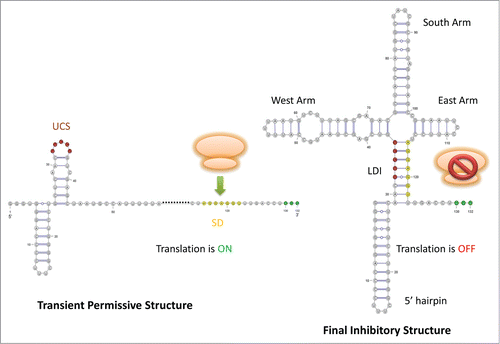
Figure 3. Schematic drawing for HDV ribozyme. The starting point of each structure on the viral genome in relation to the self-cleavage site (this site is annotated by an orange arrow) is labeled by the corresponding number. The 5 stems of the active structure are colored using similar color scheme employed by Chadalavada et al. (2000) [Citation12]: P1 in blue, P2 in green, P3 in yellow, P4 in dark red, P1.1 in purple. The stem P(-1) of the permissive alternative structure is colored in pink. In the inhibitory alternative structure, Alt 1, 2, and 3 disrupt the native stems of the active structure except P1 and P4.
![Figure 3. Schematic drawing for HDV ribozyme. The starting point of each structure on the viral genome in relation to the self-cleavage site (this site is annotated by an orange arrow) is labeled by the corresponding number. The 5 stems of the active structure are colored using similar color scheme employed by Chadalavada et al. (2000) [Citation12]: P1 in blue, P2 in green, P3 in yellow, P4 in dark red, P1.1 in purple. The stem P(-1) of the permissive alternative structure is colored in pink. In the inhibitory alternative structure, Alt 1, 2, and 3 disrupt the native stems of the active structure except P1 and P4.](/cms/asset/33c53266-3210-4af0-b3e9-297b67f38d64/krnb_a_1008373_f0003_oc.gif)
Figure 4. Schematic drawing for SAM riboswitch. For both RNA structures, the stem of the terminator is colored in red, and the stem of the anti-anti-terminator is colored in blue. In the SAM-unbound structure, the anti-terminator is formed by the 3' portion of the anti-anti-terminator and the 5' portion of the terminator hairpin; thus, the terminator hair can no longer form. The pseudoknot is not shown in this drawing.
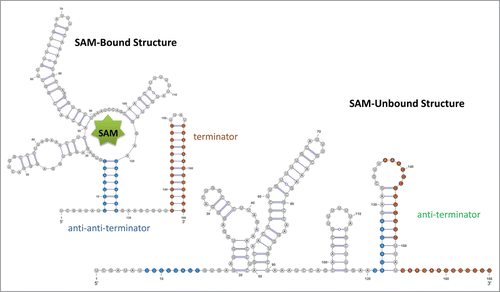
Table 1. The basic alignment statistics for our new alignments and the corresponding seed alignments in Rfam, if it exists. The structural quality measures allow a swift comparison between the structural annotation of our new alignments and that of the corresponding seed alignments in Rfam.
Table 2. The alignment quality measures for the transient structural features for our alignments. For Trp, the numbers refer to the anti-terminator structure.
Figure 5. Arc-plot of Tryptophan operon leader made using the visualisation program R-chie [Citation96]. The left legend specifies the percentage of canonical base-pairs in the paired alignment columns, i.e. those connected by an arc. The right legend specifies the evolutionary support (e.g., covariation, etc.) for each position in the alignment. The alignment and arcs at the top show the information for the terminator structure, whereas the bottom ones correspond to the anti-terminator structure for the same underlying alignment. The lines in each alignment correspond to the respective sequences with every box representing either a nucleotide or a gap in the respective sequence. Every arc represents a base-pair involving the respective two alignment columns. The arcs are colour-coded according to their percentage of canonical base pairs, whereas the evolutionary information supporting each base pair is encoded in the colouring of the underlying nucleotides in the base paired alignment columns, see the right legend for details. Two green blocks connected by an arc implies that this is a canonical base pair (i.e., GC, AU or GU) which corresponds to the most abundant type of base pair for this pair of alignment columns. Cyan means this is a canonical base pair but that it has an one-sided mutation with respect to the most abundant (green) base-pair. Blue refers to a canonical base-pair which differs on both sides from the most abundant (green) base-pair. Red means this is a non-canonical base pair. Unpaired nucleotides are shown in black and gaps in grey.
![Figure 5. Arc-plot of Tryptophan operon leader made using the visualisation program R-chie [Citation96]. The left legend specifies the percentage of canonical base-pairs in the paired alignment columns, i.e. those connected by an arc. The right legend specifies the evolutionary support (e.g., covariation, etc.) for each position in the alignment. The alignment and arcs at the top show the information for the terminator structure, whereas the bottom ones correspond to the anti-terminator structure for the same underlying alignment. The lines in each alignment correspond to the respective sequences with every box representing either a nucleotide or a gap in the respective sequence. Every arc represents a base-pair involving the respective two alignment columns. The arcs are colour-coded according to their percentage of canonical base pairs, whereas the evolutionary information supporting each base pair is encoded in the colouring of the underlying nucleotides in the base paired alignment columns, see the right legend for details. Two green blocks connected by an arc implies that this is a canonical base pair (i.e., GC, AU or GU) which corresponds to the most abundant type of base pair for this pair of alignment columns. Cyan means this is a canonical base pair but that it has an one-sided mutation with respect to the most abundant (green) base-pair. Blue refers to a canonical base-pair which differs on both sides from the most abundant (green) base-pair. Red means this is a non-canonical base pair. Unpaired nucleotides are shown in black and gaps in grey.](/cms/asset/ddabdbbd-a9cc-4517-aefe-db703ca77f40/krnb_a_1008373_f0005_oc.gif)
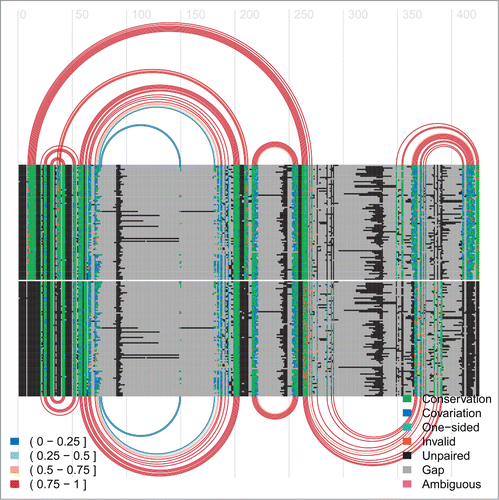
Figure 6. Arc-plot for the Levivirus alignment. The alignment and arcs at the top correspond to the final inhibitory structure, whereas the bottom ones correspond to the transient structure permissive for maturation protein translation. See the caption of for more information on arc-plots and two legends.
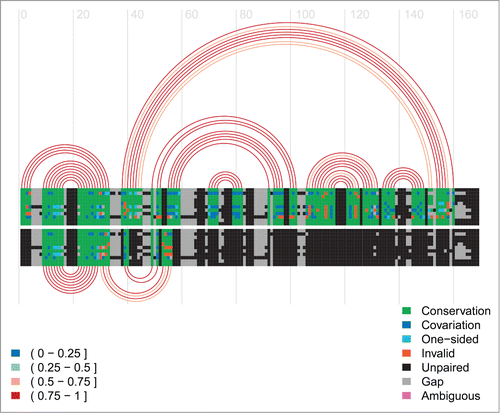
Figure 7. Arc-plot for the HDV ribozyme. The alignment and arcs at the top show the permissive alternative structure 2 and the active structure. The alternative structure 2, P(-1), is the 13-bp hairpin on the left side on top of the arc diagram. The bottom arcs correspond to the inhibitory alternative structure 1. See the caption of for more information on arc-plots and two legends.
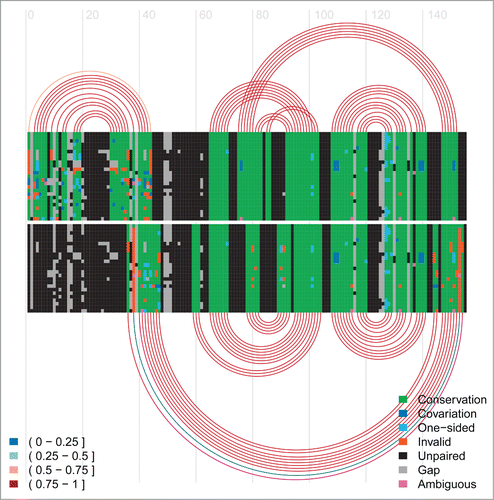
Figure 8. Arc-plot of SAM riboswitch. The top covariation and arcs correspond to the SAM-bound conformation and the bottom ones refer to the SAM-unbound conformation.