Figures & data
Figure 1. Critical events in early spliceosome assembly steps. U1 snRNP interacts with the functional 5' splice site (5'ss), SF1 recognizes branchpoint sequence (BPS), and U2 snRNP auxiliary factors bind the polypyrimidine tract (Py-tract) between the BPS and AG dinucleotide. After U2 snRNP binding at the 3' splice site (3'ss), SF1 and U2AF heterodimer detach from the spliceosome followed by the joining of the U4/U6.U5 tri-snRNPs in an ATP-dependent manner to convert pre-spliceosome to mature spliceosome.
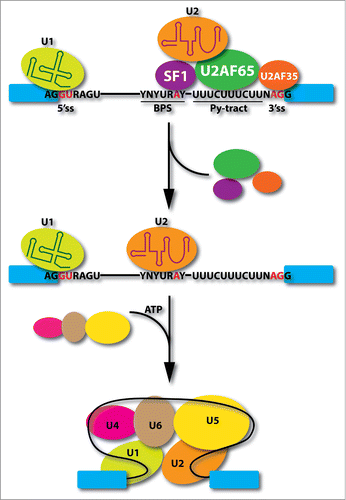
Figure 2. Domain structures of human U2AF-related proteins. (A), Human U2AF65 (NCBI Accession No. NP_009210) and its paralogues RBM39/CAPERα (NP_004893), RBM23/CAPERβ (NP_060577) and PUF60 (NP_001258027). (B), Human U2AF35 (NP_006749) and its paralogues U2AF26 (NP_659424), U2AF35R1 (Q15695) and Urp/U2AF35R2 (NP_005080). RRM, RNA recognition motif (blue); UHM, U2AF homology motif (green); RS, arginine-serine-rich (deep pink); Zn, zinc knuckle (gold).
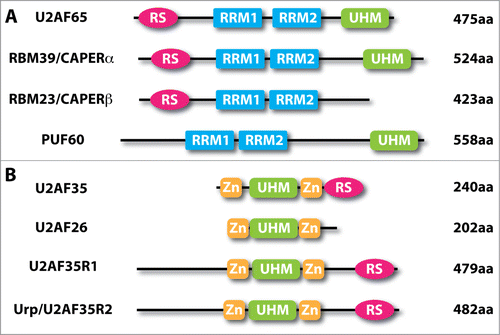
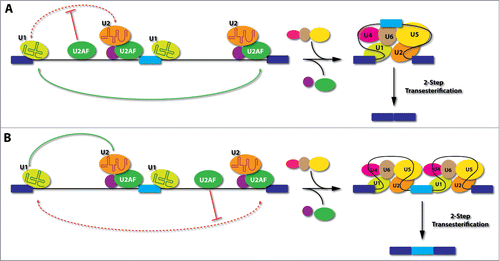
Figure 3. Model for positional-dependent effects of U2AF in the regulation of alternative splicing. (A), If U2AF binds to an intronic location upstream of the alternative exon, it will interfere with the communication between the splicing complexes assembled on the upstream 5'ss and those formed on the 3'ss of the alternative exon (upstream 3'ss). This would provide competitive advantage in selecting the 3'ss of the downstream intron (downstream 3'ss), thus causing skipping of the alternative exon. (B), If U2AF binds to an intronic location downstream of the alternative exon, it will interfere with the recognition or pairing of the downstream 3'ss, thus offering certain advantage for the upstream 3'ss to engage in productive communication with splicing complexes assembled on the upstream 5'ss. This would favor the inclusion of the alternative exon. Light blue: alternative exon; Dark blue: constitutive exons; Red dashed lines: interference of splice site pairing; Green lines: preferred pairing.