Figures & data
Figure 1. Examples of tRNA modifications found in the ASL. Enzyme-catalyzed modification of tRNA can promote anticodon-codon interactions, with modifications at positions 34 and 37 having mechanistic involvement in disease pathologies and stress signaling.
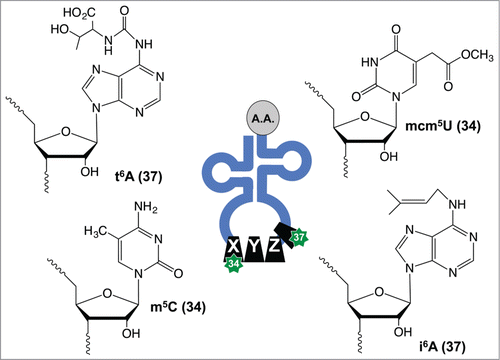
Figure 2. Alteration of tRNA structure promotes functional regulation. Regulation of tRNA transcription, processing and modification has been reported in response to many stresses, with each regulatory change having the potential to contribute to changes in global tRNA modification levels. Reprogramming of tRNA modifications has been linked to tumor suppression, mitochondrial diseases, cell cycle progression and DNA damage- and ROS-responses, among other stress response pathways.
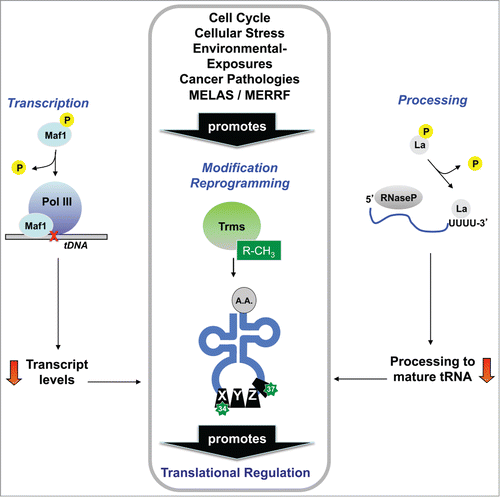
Figure 3. A platform for systems-level quantification of stress-induced changes in tRNA modifications links them to regulation of specific genes based on codon usage. The pipeline for identifying connections between specific tRNA modifications and codon-biased genes begins with (1) exposing cells to different stresses, (2) isolation and hydrolysis of tRNA, (3) HPLC resolution of individual ribonucleosides, followed by quantification of each ribonucleoside by mass spectrometry, (4) analysis of stress-induced changes by multivariate statistical analysis, (5) assignment of significantly altered ribonucleosides to specific tRNAs, and (6) analysis of the cognate codons in genome-wide, gene-specific methods to identify codon trends in stress-response transcripts.
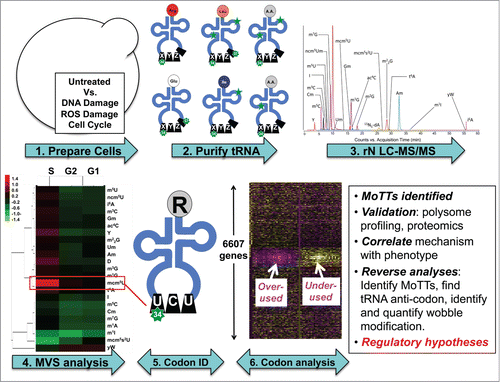
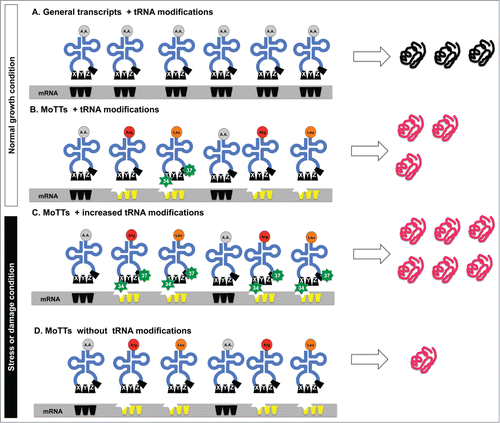
Figure 4. Model for the regulation of MoTTs and ordinary transcripts under normal and stress conditions. Different stress conditions promote reprogramming of tRNA modifications that affects regular transcripts and MoTTs differentially. Under normal conditions, both (A) regular transcripts that are not codon biased and (B) MoTTs are efficiently translated. However, in response to stress-induced reprogramming of tRNA modifications, (C) MoTTs have their translation stimulated because the codon-bias (represented as indented stars) can be decoded by the stress-specific ASL modifications (green stars). In the absence of modified tRNA (D), MoTTs are poorly translated and this leads to decreased levels of stress response proteins and sensitivity to the stress.