Figures & data
Table 1. Preferences for the different RNA conformations in RNA interfaces bound to different RNA-protein domainsFootnote*
Figure 1. Clustering the RNAs from all protein-RNA complexes according to their different conformations and sequence features. The conformations and sequences of the RNAs extracted from all protein-RNA complexes were analyzed and represented as feature vectors. The vectors representing either the conformation or sequence signatures of the RNA from each complex were further clustered independently to 4 distinct clusters, employing the K-means clustering. (A) A heatmap representing the average frequency of each RNA conformation for all RNAs in each of the 4 clusters (B) The distribution of domain families within each of the 4 clusters that were classified according to the RNA conformations (C) A heatmap representing the average frequency of each of the 4 nucleobases in all RNA sequences within each of the 4 clusters (D) The distribution of domain families within each of the 4 clusters that were classified according to the nucleotide composition.
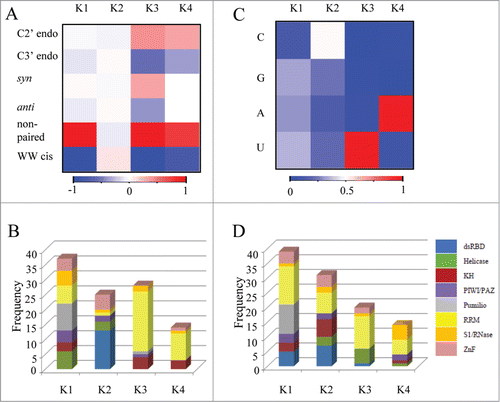
Figure 2. The distribution of Syn and C2' endo conformations in dinucleotides binding to the RRM domain. Asterix indicates nucleotides in rare conformation (syn or C2' endo) (A) Heatmap demonstrating the statistical significant of the enrichment of the dinucleotides possessing the syn conformations in RNA bound to RRM relative to the background of All RNA. The statistical significance is represented by the –log P value of the enrichment calculated using the Fisher's exact test following the Bonferroni correction, color range from azure (not significant) to blue (highly significant) (B) Heatmap demonstrating enrichment of the dinucleotides possessing the C2' endo conformations in RNA bound to RRM relative to the background of All RNA. The statistical significance is represented by the –log P value calculated using the Fisher's exact test following the Bonferroni correction, color range from azure (not significant) to blue (highly significant).
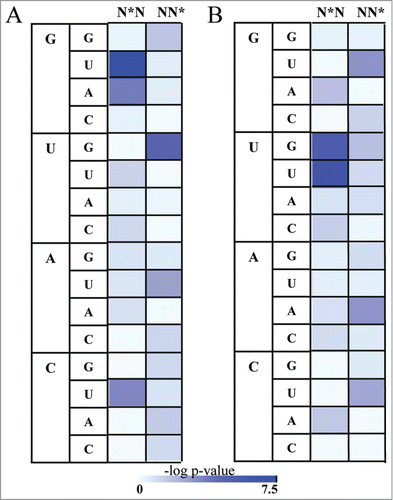
Figure 3. The preference of syn and C2' endo conformation in RRM-RNA interactions. Nucleotides in syn conformation or C2' endo conformation are presented as following: A are presented as red, U are presented as green, C are presented as blue and G are presented as yellow. Nucleotides in anti conformation or not in C2' endo conformation which are involved in the interactions are shown as gray. P value (Fisher's exact test) indicates the statistical preference of nucleotides in syn or C2' endo conformations to be involved in interactions between the RNA and the RRM domain, relative to the general abundance in all other protein-RNA interactions.
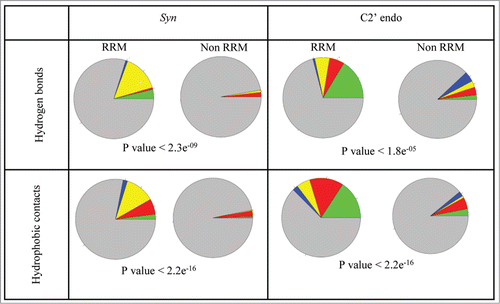
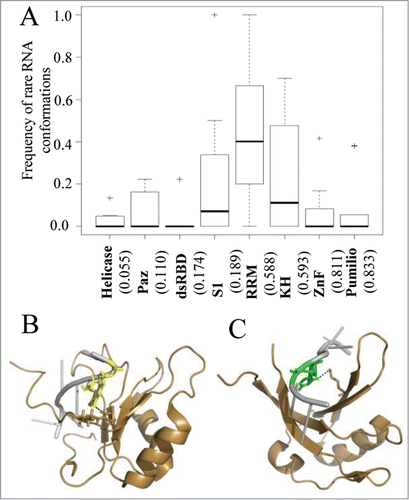
Figure 4. The interplay between sequence specificity and rare RNA conformations. (A) Box plots demonstrate the frequencies of interactions with rare RNA conformations in RNAs from protein-RNA complexes grouped according the interacting RNA binding domain family. The numbers in the x-axis below each plot indicate the calculated average frequency of specific hydrogen bond interactions within each family. In (B and C) are examples of RRM-RNA complexes from the PDB which involve hydrogen bond interactions between the RRM domain and nucleotides with rare conformations. The RNA and protein are colored in silver and sand, respectively. Hydrogen bonds between the protein domain and the RNA are depicted in dashed black lines. (B) Graphical representation of the interactions of the RRM domain with nucleotide in syn conformation (PDB ID 2LEC). Highlighted are the interacting pair G (yellow) and ARG (stick). (C) Graphical representation of the interactions of the RRM domain with nucleotide in C2' endo conformation (PDB ID 2XS2). Highlighted are the interacting pair U (green) and LYS (stick).