Figures & data
Figure 1. Knockdown of Dis3 in the wing imaginal disc results in severe developmental phenotypes. Knockdown of Dis3 in the wing pouch of the wing imaginal disc results in a “no wing” phenotype at 100% penetrance (Dis3KD) (B) when compared to a UAS-Dis3RNAi parental control male fly (Dis3WT) (A). (C) Knockdown of Dis3 protein in nub-GAL4/+; UAS-Dis3RNAi/+ wing imaginal discs by Western blotting. Dis3WT genotype: ; nub-Gal4;. (C) Dis3 expression is knocked down to 20% of parental controls in wing imaginal discs when UAS-Dis3RNAi is driven by nub-GAL4. 'Dis3WT' includes parental genotypes ; nub-Gal4 ; and ;; UAS-Dis3RNAi. n ≥ 3, p = 0 .0009, error bars show standard error. (D and E) nub-GAL4/+; UAS-Dis3RNAi/+ flies also show a severe lack of haltere development (E) (100% penetrance) compared to parental control flies (D). (F) Dis3 knockdown progeny (nub-GAL4/+ ; UAS-Dis3RNAi/+) are delayed in larval development by 40 hours (red) when compared to parental controls (black – UAS-Dis3RNAi and nub-GAL4) (n ≥ 23 ).
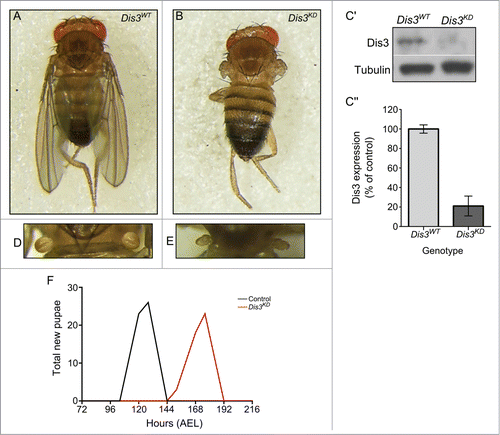
Figure 2. Dis3 knockdown in the wing pouch of the wing imaginal discs results in small wing discs and extensive apoptosis. (A and B) Dis3 knockdown (nub-GAL4/+ ; UAS-Dis3RNAi/+) late L3 wing imaginal discs (B) are smaller than parental control (Dis3WT is ;; UAS-Dis3RNAi) wing imaginal discs (A) (scale bar=50μm). (C) Quantitation of nub-GAL4 driven Dis3 knockdown wing imaginal discs shows that they are 23% smaller than parental control discs ('Dis3WT' includes parental genotypes ; nub-GAL4 ; and ;;UAS-Dis3RNAi). (n ≥ 21, P < 0.0001, error bars represent standard error). (D and E) Knockdown of Dis3 in the wing pouch results in apoptosis specifically localized within the wing pouch region (E) while parental control discs show very little apoptotic activity (D). (n ≥ 26, scale bar = 50 μm).
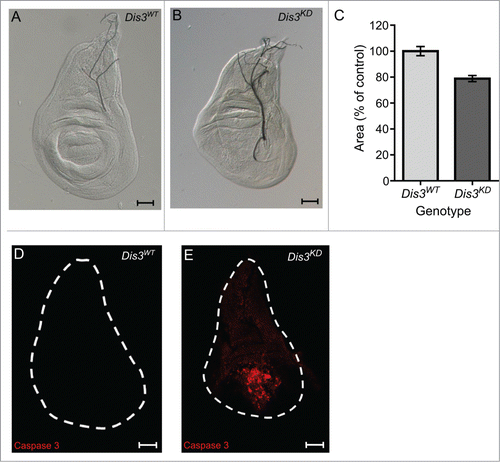
Table 1. Stages of lethality observed following ubiquitous knockdown of Dis3
Figure 3. (See previous page). Expression patterns of miRNA in wing imaginal discs. (A) Kernel density plot of the general distribution of the 109 detected miRNAs indicates 2 peaks of miRNA expression. Knockdown of Dis3 results in a shift in the poorly expressed miRNAs (left hand peak) to a higher level of expression compared to the more highly expressed miRNAs which do not change (right hand peak). (B) Distribution of fold changes between both parents (Control) and grouped parents vs Dis3 knockdown (Dis3KD). Knockdown of Dis3 results in the emergence of peaks around +/− 2-fold change. Dotted vertical lines represent +/− 2-fold change. (C) Comparison of miRNA RPKMs between the 2 parental controls shows high similarity. Dotted lines show +/− 2-fold change. (D) Comparison of miRNA RPKM between grouped parental controls (Control) and Dis3 knockdown (Dis3KD) wing imaginal discs. Blue and red dotted lines represent +2-fold change and −2-fold change respectively. Selected miRNAs that increase or decrease in expression following Dis3 knockdown are colored in blue and red respectively. Selected miRNAs that remain unchanged are highlighted in green. (E) 61.5% of miRNAs in the wing imaginal disc do not change in expression following the knockdown of Dis3. 26.6% of miRNAs increase in expression >1.5-fold (10.1% ≥ 2-fold). 11.9% of miRNAs decrease in expression >1.5-fold (5.5% ≥ 2-fold).
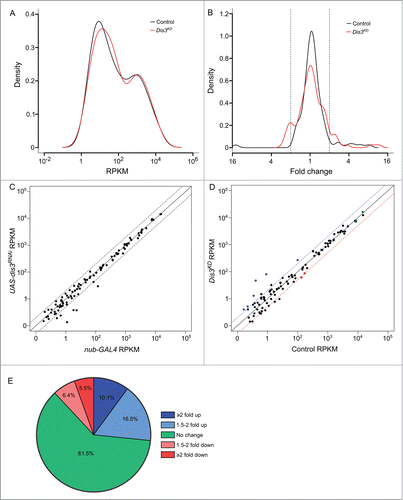
Figure 4. Effect of Dis3 knockdown on all wing imaginal disc miRNAs detected above threshold levels. Fold changes (Dis3KD vs grouped parental controls) from miRNA-seq data of the 109 detected miRNAs in the wing imaginal disc. miR-987–5p shows the greatest increase in expression (49.5-fold) while miR-125–5p shows the greatest decrease in expression (−2.4-fold). miRNAs that increase ≥2-fold are highlighted in blue font and those selected for further analysis are shown with blue bars. miRNAs that decrease ≥2-fold are highlighted in red font and those selected for further analysis shown with red bars. Selected miRNAs that remain unchanged are highlighted in green font. Dotted lines represent +/−2-fold changes. (n ≥2, error bars represent standard error).
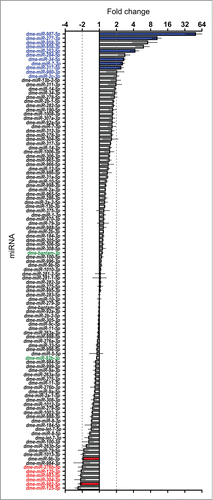
Figure 5. Identification of a novel mature miRNA in the wing imaginal disc. Using miRDeep2 we identified a novel miRNA of moderate expression in the wing imaginal discs. The predicted pre-miRNA forms a hairpin loop secondary structure with the novel miRNA being situated at the 3' end (shown in pink). The novel miRNA was detected at moderate levels using miRNA-seq in all but one sample. Validation of the miRNA using qRT-PCR shows that this novel miRNA is expressed at similar levels to miR-277–3p in all samples.
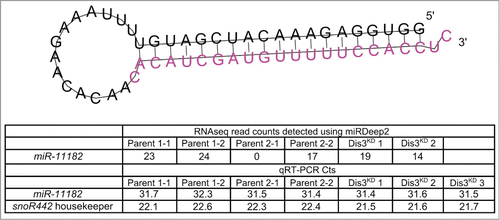
Figure 6. qRT-PCR validation of missexpressed miRNAs from the miRNA sequencing data. (A) Grouping of RPKM of selected miRNAs for all replicates is highly consistent between controls (black dots) and knockdown replicates (red dots). (B) Validation of miRNA-seq fold changes (red dots) using qRT-PCR. All selected miRNAs that change in expression upon Dis3-depletion in the miRNA-seq data also significantly change in expression when using qRT-PCR (except miR-7–3p). Dotted lines show +/−2-fold changes. Stars represent levels of significance calculated using a 2 sample t-test. (****p < 0.0001, **p < 0.01, *p < 0.05 n ≥ 3, error bars represent standard error). (C) The pri/pre-miRNA levels were measured for the miRNAs that significantly change in expression in both the miRNA-seq and qRT-PCR data. The expression levels of miR-252–5p and miR-982–5p change post-transcriptionally following Dis3 knockdown. All other miRNAs alter in a transcriptional manner as the pri/pre-mRNA (light gray) also changes significantly in expression, similar to the levels observed mature miRNAs (dark gray). Dotted lines show +/−2-fold changes. Stars represent levels of significance calculated using a 2 sample t-test. ****p < 0.0001, **p < 0.01, *p < 0.05, ns p > 0.05. (n ≥ 3, error bars represent standard error).
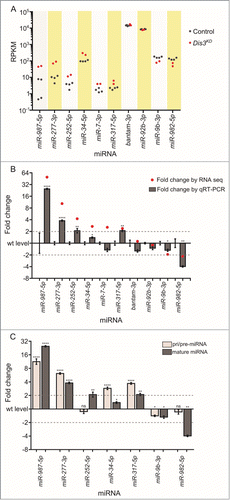
Figure 7. Overexpression of miR-252 in the wing pouch of the imaginal disc results in severe wing phenotypes. (A) Overexpression of miR-252 using the ubiquitously expressed tub-GAL4 driver results in 100% lethality compared to controls (UAS-miR-252/TM6). n = 184. (B) Overexpression of UAS-miR-252 in the wing pouch using the nubbin-GAL4 driver results in a 1154-fold increase in miR-252–5p expression in the wing imaginal disc. (n ≥ 3, p < 0.0001, error bars represent standard error). (C and D) Overexpression of UAS-miR-252 in the wing pouch of the wing imaginal disc using the nubbin-GAL4 driver (;nub-Gal4/+ ;UAS-miR-252/+) results in severe wing crumpling (D) compared to a parental control (miR-252WT is UAS-miR-252) (C) with 100% penetrance. n = 168.
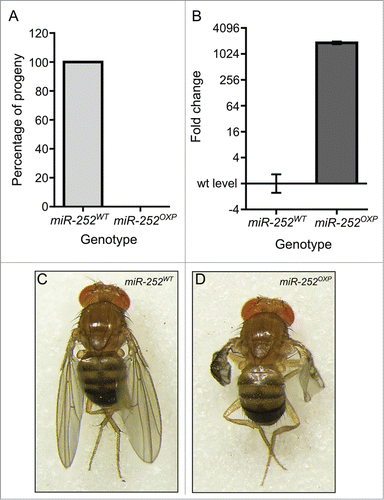
Figure 8. Knockdown of the exosome subunit Rrp40 results in similar phenotypes as to Dis3 knockdown. Knockdown of the exosome subunit Rrp40 using the nubbin-GAL4 wing pouch driver (;nub-Gal4/UAS-Rrp40RNAi;) results in severe wing development phenotypes (B' and B”) when compared to parental controls (A). 90% of adults present a severe phenotype with crumpled wings (B') and the remaining 10% show a similar phenotype to Dis3 knockdown with no wings (B”). (C) Knockdown of Rrp40 in the wing pouch (nub-GAL4) results in a similar increase in miR-252–5p expression as observed during Dis3 depletion, indicating the effect observed upon Dis3 knockdown is a result of Dis3/exosome impairment (n≥3, **p < 0.01, error bars represent standard error).
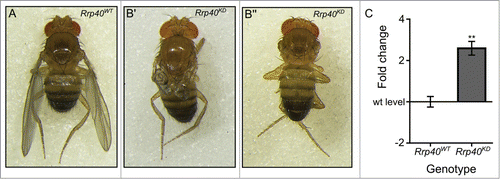
Table 2. Summary of missexpressed miRNAs in Dis3 knockdown wing imaginal discs
