Figures & data
Figure 1. Production of HER2 variants. (A) A regulation hotspot in HER2. The region between exon 14 and exon 17 produces of a number of different HER2 variants. Including Δ16HER2 which has exon 16 excluded, p100 which has intron 15 included, and X5 which splices in part of intron 14. In addition to these splice variants there are 2 further variants produced by alternative translation initiation in exon 15 and 17 (CTF-611 and CTF-687). (B) The other splice variant of HER2 (Herstatin) is produced by the inclusion of intron 8.
Table 1. siRNA sequences
Table 2. Primer sequences
Table 3. Oligonucleotide sequences for RNase-assisted chromatography
Figure 2. HER2 mRNA expression in breast tumor tissue and SKBR3 breast cancer cells. (A) HER2 mRNA expression was investigated in a cohort of 12 breast tumor tissue samples and the HER2 positive SKBR3 breast cancer cell line. Splice-specific primers for exon16 inclusion (WT), exon16 exclusion (Δ16HER2), intron 15 inclusion (p100) and exon 15a inclusion (X5) were used to determine expression of HER2 splice variants along with 2 housekeeping genes GAPDH and β-actin. (B) Intracellular expression of WT HER2 mRNA in live SKBR3 breast cancer cells using SmartFlare™ RNA detection probes.
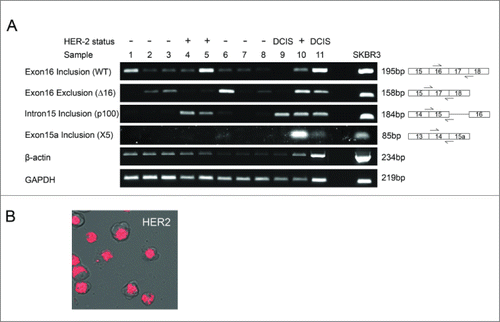
Figure 3. HER2 proteins expressed in breast tumor tissues and SKBR3 cells. (A and B) Reduction of HER2 protein expressed by SKBR3 cells was achieved via RNA interference using 2 different siRNA sequences (lanes 1–4). Four controls comprising a positive transfection control (GAPDH siRNA (G)), a negative scrambled sequence control siRNA (NC), a vehicle only control (V) and untreated cells (UT) were included (lanes 5–8). Levels of HER2 proteins were determined by immunoblotting using C-terminal (A) and N-terminal (B) anti-HER2 antibodies (C) Immunoblotting using C-terminal and N-terminal anti-HER2 antibodies and protein extracted from tumor tissue samples and SKBR3 cells. GAPDH was used as a loading control.
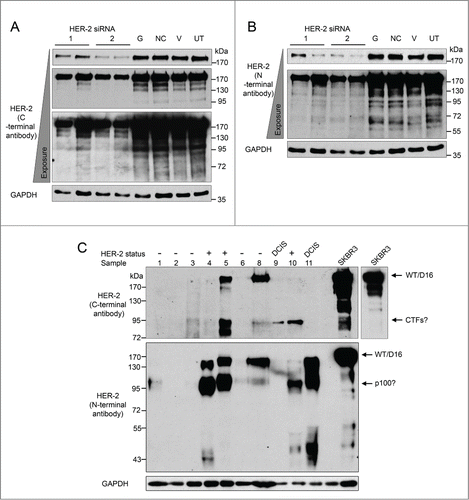
Figure 4. Effect of the splicing factor SRSF3 on HER2 mRNA transcripts in SKBR3 cells. RNA interference was used to reduce the levels of SRSF3 in SKBR3 cells (SRSF3 (1)). Four controls comprising a positive transfection control (GAPDH siRNA (G)), a negative scrambled sequence control siRNA (NC), a vehicle only control (V) and untreated cells (UT) were also used. Reduction of SRSF3 mRNA was determined at 48 hours by classical PCR (A) and reduction of SRSF3 protein was determined by immunoblotting at 72 hours after transfection (B). (C) Alternative splicing events were measured using splice specific primers by qPCR at 72 hours post-transfection. (mean (n = 3) ± SEM). * P ≤ 0.05, *** P ≤ 0.001.
Figure 5 . Screening splicing factors for effects on HER2 mRNA transcripts in SKBR3 cells. RNA interference was used to reduce the levels of splicing factors (SRSF1, SRSF2, SRSF5, hnRNP A1, hnRNP H1, and YB1) in SKBR3 cells. Four controls comprising a positive transfection control (GAPDH siRNA (G)), a negative scrambled sequence control siRNA (NC), a vehicle only control (V) and untreated cells (UT) were also used. (A) Reduction of mRNA was determined at 48 hours by classical PCR. (B) HER2 alternative splicing events were measured using splice-specific primers by qPCR at 72 hours post-transfection (mean (n = 3) ± SEM). * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001.
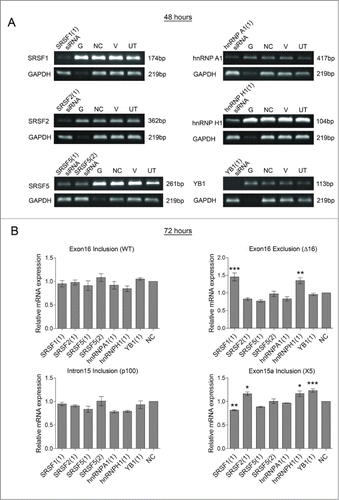
Figure 6. Confirming effect of splicing factors on HER2 mRNA transcripts in SKBR3 cells. A second siRNA sequence was used to reduce the levels of splicing factors (SRSF1, SRSF2, hnRNP H1, and YB1) in SKBR3 cells. Four controls comprising a positive transfection control (GAPDH siRNA (G)), a negative scrambled sequence control siRNA (NC), a vehicle only control (V) and untreated cells (UT) were also used. (A) Reduction of mRNA was determined at 48 hours by classical PCR. (B) HER2 alternative splicing events were measured using splice-specific primers by qPCR at 72 hours post-transfection (mean (n = 3) ± SEM). * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001.
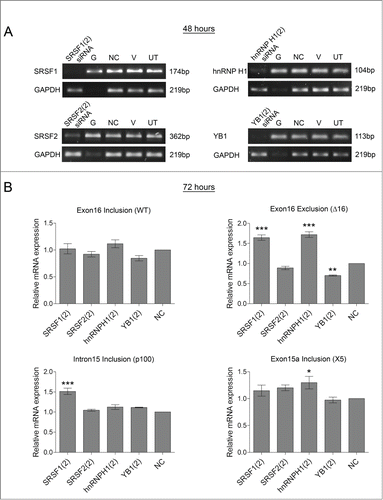
Figure 7. Identification of RNA binding proteins bound to the splicing hotspot of HER2. (A) Binding of splicing factors to endogenous HER2 mRNA (exons exon16-exon17/18) and pre-mRNA (exon11-intron11) was determined by RNA immunoprecipitation using antibodies that target SRSF3 and hnRNP H1. Fold enrichment of target mRNA was determined by qPCR. (B) DNA oligonucleotides were designed to cover the regions surrounding exon 15 (B) and exon 16 (C). A further oligonucleotide was designed which is identical to oligonucleotide H3 apart from having 2 potential SRSF3 binding sites mutated. RNase-assisted RNA chromatography was carried out on RNA produced by the oligonucleotides along with a control (CT) where no RNA was added to the beads. Immunoblotting was performed with the RNase treated supernatant using antibodies against SRSF3, and hnRNP H1 and nuclear protein extract (NE) was included as a positive control.
